A Causality-Guided KAN Attention Framework for Brain Tumor Classification
-
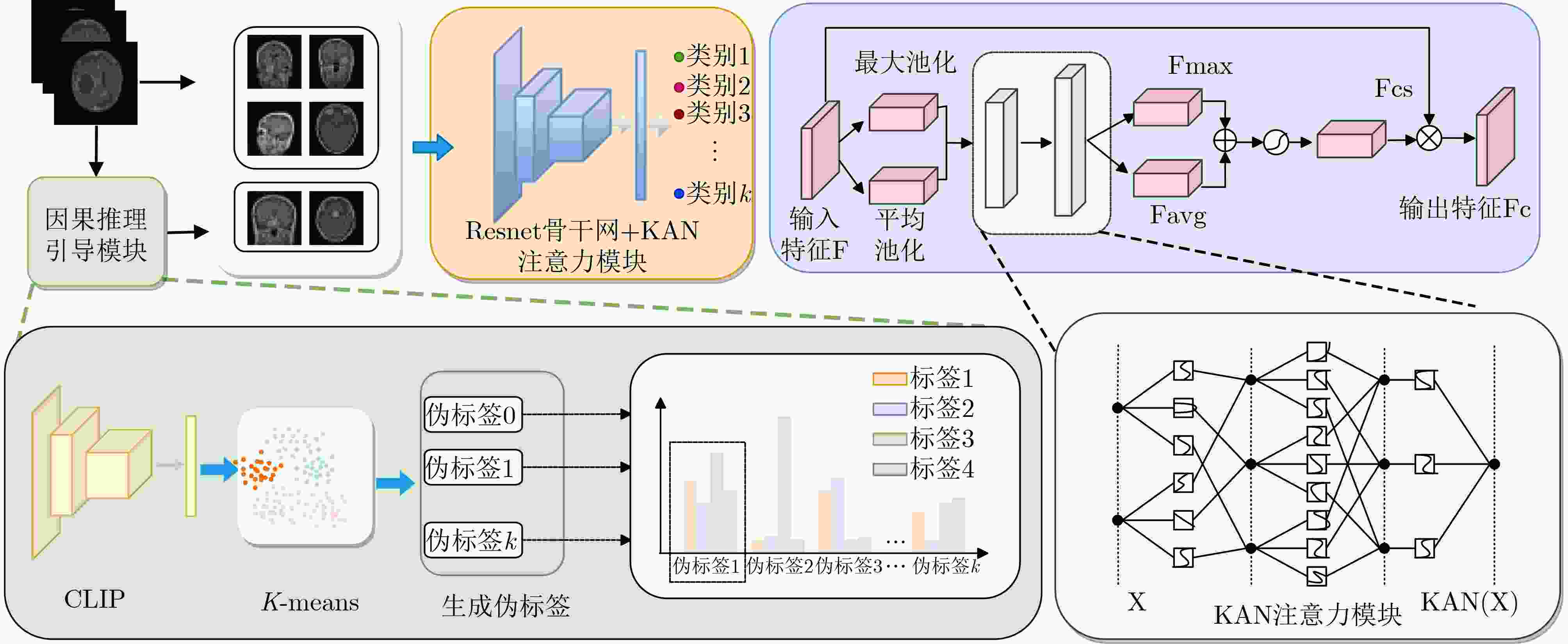
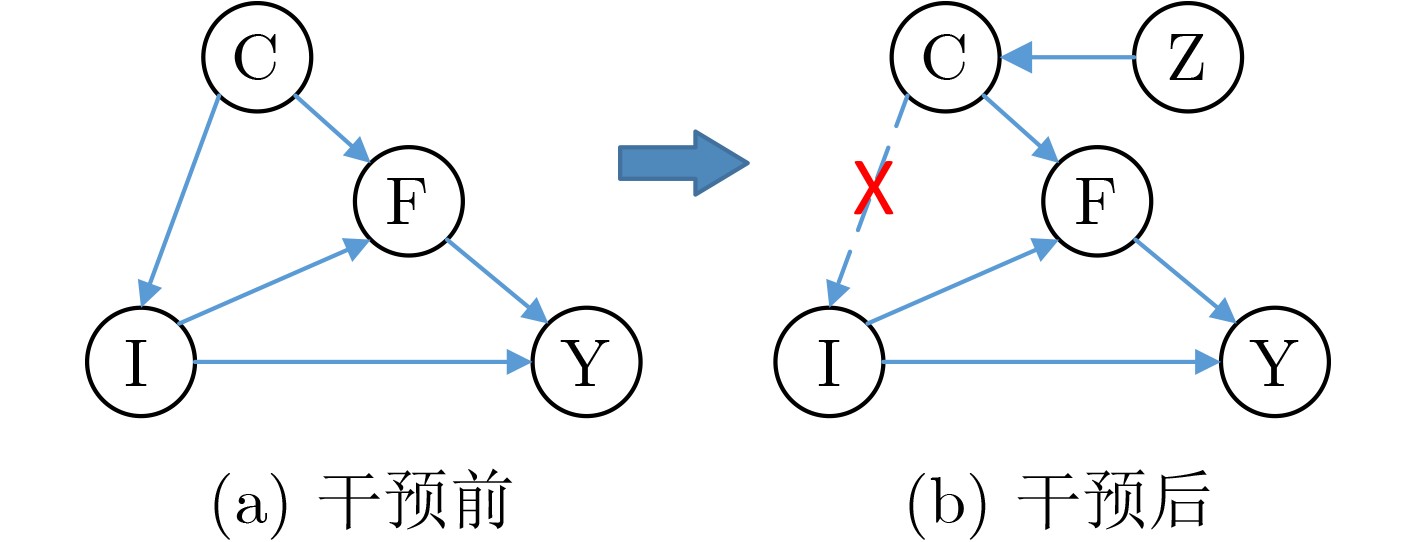
摘要: 脑肿瘤分类是医学影像分析中的关键任务,但现有深度学习方法在应对扫描参数差异、解剖位置偏移等因素时仍面临特征混淆问题,且难以建模肿瘤异质性引发的复杂非线性关系。针对这一挑战,本文提出一种因果推理引导的KAN注意力分类框架。首先,基于CLIP模型进行无监督特征提取,捕捉MRI数据中的高层语义特征;其次,基于K-means聚类设计混淆均衡度指标,筛选混淆因子图像。并设计因果干预机制,显式引入混淆样本,同时提出因果增强的损失函数以优化模型的判别能力;最后,在预训练ResNet主干网中引入KAN注意力模块,强化模型对肿瘤局部坏死区与强化边缘的非线性关联建模能力。实验表明,所提出的方法在脑肿瘤分类任务中优于传统CNN与Transformer模型,验证了其在判别能力和鲁棒性方面的优势。本研究为医学影像的因果推理与高阶非线性建模提供了新的技术路径。Abstract:
Objective In recent years, Convolutional Neural Network (CNN)-based Computer-Aided Diagnosis (CAD) systems have advanced brain tumor classification. However, classification performance remains limited due to feature confusion and inadequate modeling of high-order interactions. To address these challenges, this study proposes an innovative framework that integrates causal feature guidance with a KAN attention mechanism. A novel metric, the Confusion Balance Index (CBI), is introduced to quantify real label distribution within clusters. Furthermore, a causal intervention mechanism is designed to explicitly incorporate confused samples, enhancing the model’s ability to distinguish causal variables from confounding factors. In addition, a KAN attention module based on spline functions is constructed to accurately model high-order feature interactions, thereby strengthening the model’s focus on critical lesion regions and discriminative features. This dual-path optimization approach, combining causal modeling with nonlinear interaction enhancement, improves classification robustness and overcomes the limitations of traditional architectures in capturing complex pathological feature relationships. Methods This study employs a pre-trained CLIP model for feature extraction, leveraging its representation capabilities to obtain semantically rich visual features. Subsequently, based on K-means clustering, the Confusion Balance Index (CBI) is introduced to identify confusing factor images, and a causal intervention mechanism is implemented to explicitly incorporate these confused samples into the training set. Based on this, a causal-enhanced loss function is designed to optimize the model’s ability to discriminate causal variables from confounding factors. Furthermore, to address insufficient high-order feature modeling, a Kolmogorov-Arnold Network (KAN)-based attention mechanism is further integrated. This module, built on spline functions, constructs flexible nonlinear attention representations to finely model high-order feature interactions. By fusing this module with the backbone network, the model achieves enhanced discriminative performance and generalization capabilities. Results and Discussions The proposed method significantly outperforms existing approaches across three datasets. On DS1, the model achieves 99.92% accuracy, 99.98% specificity, and 99.92% precision, surpassing benchmarks such as RanMerFormer (+0.15%) and SAlexNet (+0.23%), with improvements exceeding 2% over traditional CNN methods (95%–97%). Although Swin Transformers achieve 98.08% accuracy, their precision (91.75%) highlights the superior robustness of our method in reducing false detections. On DS2, the model attains 98.86% accuracy and 98.80% precision, outperforming the second-best RanMerFormer. On a more challenging in-house dataset, the model maintains 90.91% accuracy and 95.45% specificity, demonstrating generalization to complex scenarios. Performance gains are attributed to the KAN attention mechanism's high-order feature interaction modeling and the causal reasoning module's confounding factor decoupling. These components enhance focus on critical lesion regions and improve decision-making stability in complex scenarios. Experimental results validate the framework's superiority in brain tumor classification, offering reliable support for clinical precision diagnostics. Conclusions Experimental results substantiate that the proposed framework confers marked improvements in brain tumor classification, with the synergistic interaction between the causal intervention mechanism and the KAN attention module serving as the principal factor driving performance gains. Notably, these enhancements are achieved with negligible increases in model parameters and inference latency, thereby ensuring both efficiency and practicality. This study delineates a novel methodological paradigm for medical image classification and underscores its prospective utility in few-shot learning scenarios and clinical decision support systems. -
表 1 数据集信息
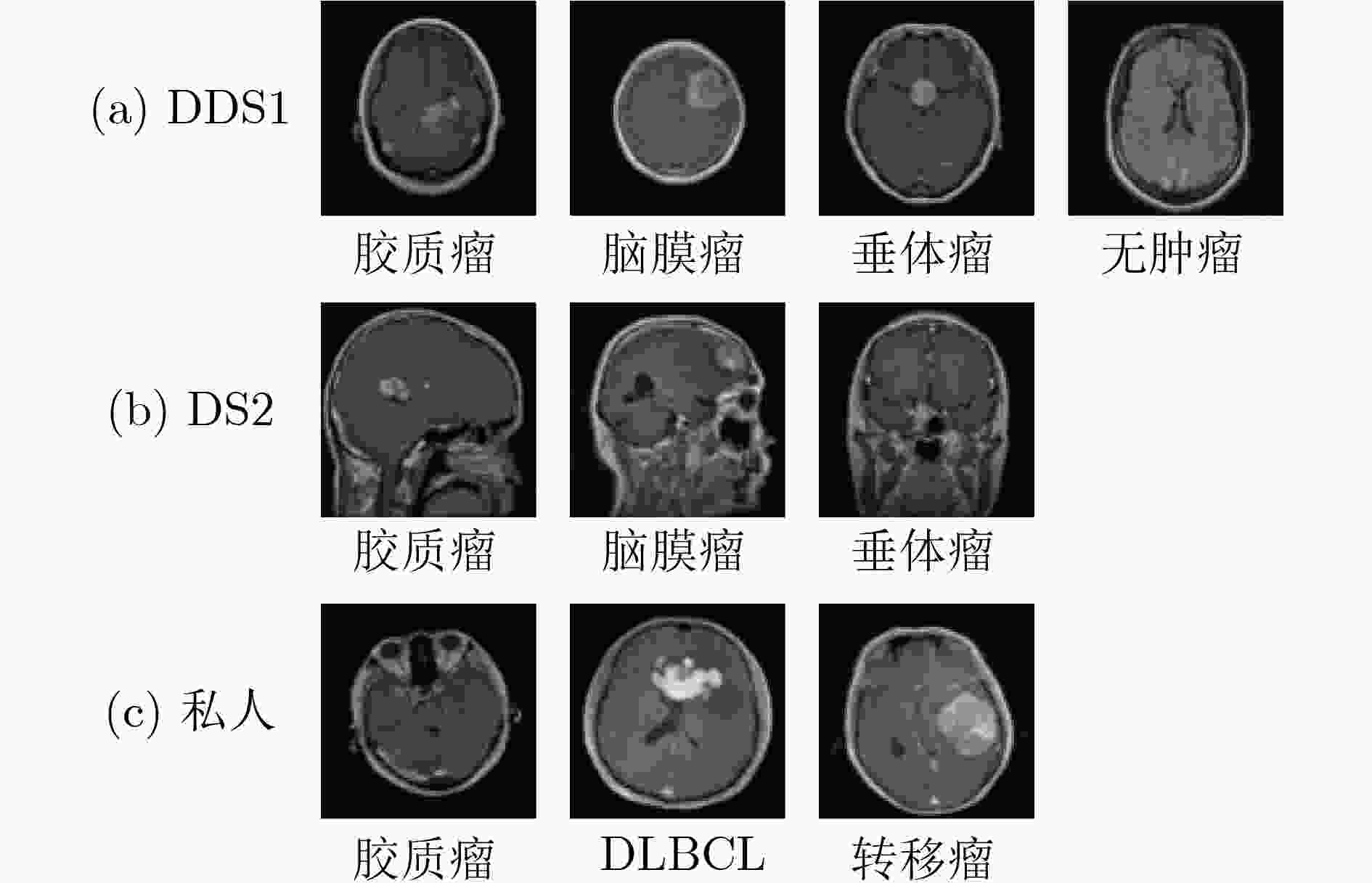
肿瘤类型 DS1 DS2 私人 脑膜瘤 1645 708 - 垂体瘤 1757 930 - 胶质瘤 1621 1426 891 DLBCL - - 468 转移瘤 - - 743 无肿瘤 2000 - - 总 数 7023 3064 2102 训练集 5712 2451 1680 测试集 1311 613 422 表 2 不同模型的测试集结果比较(%)
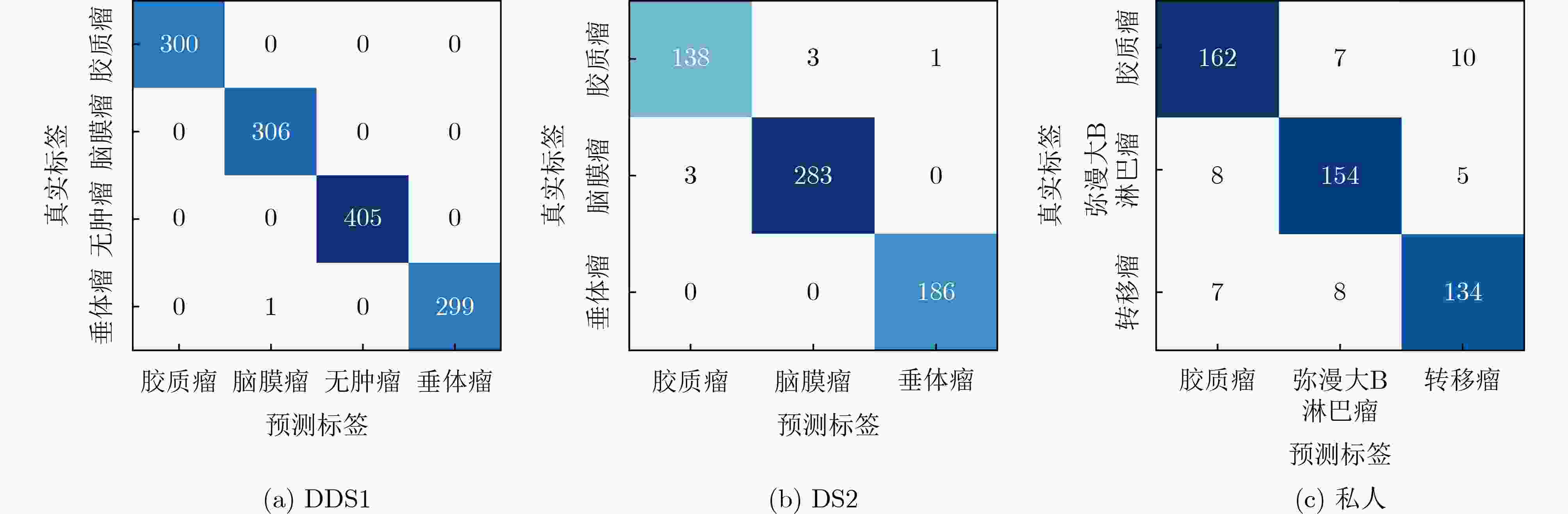
数据集 方法 Pr Se Sp Acc DS1 FTVT[24] 98.71 98.70 - 98.70 Swin Transformer V2[25] 98.75 98.51 - 98.97 NeuroNet19[26] 99.20 99.20 - 99.30 ResVit[27] 98.45 98.61 - 98.47 InceptionV3[28] 97.97 96.59 99.98 97.13 CNN[29] 97.00 97.00 - 97.00 DCST+SVM [30] 97.80 96.62 - 97.71 Swin Transformers [13] 91.75 - - 98.08 Custom built CNN[31] 95.00 95.00 - 95.16 RanMerFormer [10] 99.76 99.75 99.93 99.77 SAlexNet[9] 99.37 99.33 - 99.69 TinyViT*[41] 99.43 99.44 99.83 99.47 LeViT*[42] 98.92 98.93 99.68 99.01 Mobilenet-v4*[43] 97.99 97.98 99.37 98.09 Ours 99.92 99.92 99.98 99.92 DS2 ARM-Net[32] 96.46 96.09 - 96.64 GT-Net[33] - - - 97.11 ResVit[27] 98.54 98.54 - 98.53 GAN+ConvNet[34] 95.29 94.91 97.69 95.60 CNN+SVM[35] 97.30 97.60 98.97 98.00 RanMerFormer[10] 98.87 98.46 99.39 98.86 Gaussian CNN[36] 97.07 - - 97.82 DACBT[37] - 98.09 100 98.56 Custom built CNN[11] 96.06 94.43 96.93 96.13 VGG19+CNN[38] 98.34 98.60 99.28 98.54 GoogleNet[39] 97.20 97.30 98.96 97.10 TinyViT*[41] 97.73 98.01 99.02 98.05 LeViT*[42] 97.10 97.07 98.68 97.39 Mobilenet-v4*[43] 97.45 97.19 98.70 97.56 Ours 98.80 98.70 99.40 98.86 Ours VIT*[40] 80.88 80.85 90.60 81.21 GoogleNet*[39] 90.09 90.29 95.10 90.10 TinyViT*[41] 88.73 88.33 94.20 88.48 LeViT*[42] 80.89 80.94 90.57 81.01 Mobilenet-v4*[43] 86.72 86.72 93.35 86.46 Ours 90.86 90.88 95.45 90.91 “*”表示在统一实验设置下复现得到的结果 表 3 消融实验结果比较(%)
数据集 方法 Pr Se Sp Acc DS1 ResNet18 99.84 99.83 99.95 99.85 ResNet18+Causal 99.92 99.92 99.98 99.92 ResNet18+KAM 99.92 99.92 99.98 99.92 ResNet18+Causal+KAM 99.92 99.92 99.98 99.92 DS2 ResNet18 98.29 98.13 99.15 98.37 ResNet18+Causal 98.37 98.42 99.24 98.53 ResNet18+KAM 98.41 98.48 99.23 98.53 ResNet18+Causal+KAM 98.76 98.71 99.41 98.86 Ours ResNet18 87.28 87.46 93.66 87.27 ResNet18+Causal 89.58 89.72 94.89 89.70 ResNet18+KAM 89.61 89.79 94.88 89.70 ResNet18+Causal+KAM 90.86 90.88 95.45 90.91 表 4 模型性能分析 (%)
模型 平均Acc 参数量(M) FLOPs(G) ResNet18 95.15 11.178 1.8186 ResNet18+CAM 96.02 11.222 1.8190 ResNet+SE 92.05 11.222 1.8240 ResNet+CBAM 95.93 11.267 1.8240 ResNet18+KAM 96.56 11.188 1.8189 表 5 不同因果权重 $ \alpha $下的分类准确率 (%)
DS2 超参数$ \alpha $ 0.0 0.1 0.3 0.5 0.7 0.9 Acc(%) 98.37 98.37 98.70 98.70 98.86 98.37 表 6 不同数据集中胶质瘤的分类准确率(%)
测试集 DS1 DS2 私人 Brats2019 训练集 DS1 100 - 90.97 95.90 DS2 - 95.07 87.20 94.78 私人 88.48 87.61 95.53 85.56 -
[1] BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2024, 74(3): 229–263. doi: 10.3322/caac.21834. [2] MAHARJAN S, ALSADOON A, PRASAD P W C, et al. A novel enhanced softmax loss function for brain tumour detection using deep learning[J]. Journal of Neuroscience Methods, 2020, 330: 108520. doi: 10.1016/j.jneumeth.2019.108520. [3] BADŽA M M andBARJAKTAROVIĆ M Č. Classification of brain tumors from MRI images using a convolutional neural network[J]. Applied Sciences, 2020, 10(6): 1999. doi: 10.3390/app10061999. [4] 张奕涵, 柏正尧, 尤逸琳, 等. 自适应模态融合双编码器MRI脑肿瘤分割网络[J]. 中国图象图形学报, 2024, 29(3): 768–781. doi: 10.11834/jig.230275.ZHANG Yihan, BAI Zhengyao, YOU Yilin, et al. Adaptive modal fusion dual encoder MRI brain tumor segmentation network[J]. Journal of Image and Graphics, 2024, 29(3): 768–781. doi: 10.11834/jig.230275. [5] AFSHAR P, PLATANIOTIS K N, and MOHAMMADI A. Capsule networks for brain tumor classification based on MRI images and coarse tumor boundaries[C]. ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 2019: 1368–1372. doi: 10.1109/ICASSP.2019.8683759. [6] 方超伟, 李雪, 李钟毓, 等. 基于双模型交互学习的半监督医学图像分割[J]. 自动化学报, 2023, 49(4): 805–819. doi: 10.16383/j.aas.c210667.FANG Chaowei, LI Xue, LI Zhongyu, et al. Interactive dual-model learning for semi-supervised medical image segmentation[J]. Acta Automatica Sinica, 2023, 49(4): 805–819. doi: 10.16383/j.aas.c210667. [7] 贾熹滨, 郭雄, 王珞, 等. 一种迭代边界优化的医学图像小样本分割网络[J]. 自动化学报, 2024, 50(10): 1988–2001. doi: 10.16383/j.aas.c220994.JIA Xibin, GUO Xiong, WANG Luo, et al. A few-shot medical image segmentation network with iterative boundary refinement[J]. Acta Automatica Sinica, 2024, 50(10): 1988–2001. doi: 10.16383/j.aas.c220994. [8] SABOOR A, LI Jianping, Ul HAQ A, et al. DDFC: Deep learning approach for deep feature extraction and classification of brain tumors using magnetic resonance imaging in E-healthcare system[J]. Scientific Reports, 2024, 14(1): 6425. doi: 10.1038/s41598-024-56983-6. [9] CHAUDHARY Q U A, QURESHI S A, SADIQ T, et al. SAlexNet: Superimposed AlexNet using residual attention mechanism for accurate and efficient automatic primary brain tumor detection and classification[J]. Results in Engineering, 2025, 25: 104025. doi: 10.1016/j.rineng.2025.104025. [10] WANG Jian, LU Siyuan, WANG Shuihua, et al. RanMerFormer: Randomized vision transformer with token merging for brain tumor classification[J]. Neurocomputing, 2024, 573: 127216. doi: 10.1016/j.neucom.2023.127216. [11] SULTAN H H, SALEM N M, and AL-ATABANY W. Multi-classification of brain tumor images using deep neural network[J]. IEEE access, 2019, 7: 69215–69225. doi: 10.1109/ACCESS.2019.2919122. [12] DÍAZ-PERNAS F J, MARTÍNEZ-ZARZUELA M, ANTÓN-RODRÍGUEZ M, et al. A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network[J]. Healthcare, 2021, 9(2): 153. doi: 10.3390/healthcare9020153. [13] LIU Ze, LIN Yutong, CAO Yue, et al. Swin transformer: Hierarchical vision transformer using shifted windows[C]. Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 9992–10002. doi: 10.1109/ICCV48922.2021.00986. [14] 刘建明, 曹圣浩, 张志鹏. 融合视觉Mamba与自适应多尺度损失的医学图像分割[J]. 中国图象图形学报, 2026, 31(1): 335–348. doi: 10.11834/jig.250224.LIU Jianming, CAO Shenghao, ZHANG Zhipeng. Medical image segmentation with vision mamba and adaptive multiscale loss fusion[J]. Journal of Image and Graphics, 2026, 31(1): 335–348. doi: 10.11834/jig.250224. [15] 朱智勤, 孙梦薇, 齐观秋, 等. 融合频率自适应和特征变换的医学图像分割[J]. 中国图象图形学报, 2026, 31(1): 303–319. doi: 10.11834/jig.250100.ZHU Zhiqin, SUN Mengwei, QI Guanqiu, et al. Frequency adaptation and feature transformation network for medical image segmentation[J]. Journal of Image and Graphics, 2026, 31(1): 303–319. doi: 10.11834/jig.250100. [16] LIU Ziming, WANG Yixuan, VAIDYA S, et al. KAN: Kolmogorov-Arnold networks[C]. The 13th International Conference on Learning Representations, Singapore, Singapore, 2024. [17] RADFORD A, KIM J W, HALLACY C, et al. Learning transferable visual models from natural language supervision[C]. Proceedings of the 38th International Conference on Machine Learning, 2021: 8748–8763. (查阅网上资料, 未找到本条文献出版地, 请确认). [18] KANUNGO T, MOUNT D M, NETANYAHU N S, et al. An efficient k-means clustering algorithm: Analysis and implementation[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2002, 24(7): 881–892. doi: 10.1109/TPAMI.2002.1017616. [19] SHAO Feifei, LUO Yawei, ZHANG Li, et al. Improving weakly supervised object localization via causal intervention[C]. Proceedings of the 29th ACM International Conference on Multimedia, 2021: 3321–3329. doi: 10.1145/3474085.3475485. (查阅网上资料,未找到本条文献出版地,请确认). [20] 李锵, 王旭, 关欣. 一种结合三重注意力机制的双路径网络胸片疾病分类方法[J]. 电子与信息学报, 2023, 45(4): 1412–1425. doi: 10.11999/JEIT220172.LI Qiang, WANG Xu, and GUAN Xin. A dual-path network chest film disease classification method combined with a triple attention mechanism[J]. Journal of Electronics & Information Technology, 2023, 45(4): 1412–1425. doi: 10.11999/JEIT220172. [21] 孙家阔, 张荣, 郭立君, 等. 多尺度特征融合与加性注意力指导脑肿瘤MR图像分割[J]. 中国图象图形学报, 2023, 28(4): 1157–1172. doi: 10.11834/jig.211073.SUN Jiakuo, ZHANG Rong, GUO Lijun, et al. Multi-scale feature fusion and additive attention guide brain tumor MR image segmentation[J]. Journal of Image and Graphics, 2023, 28(4): 1157–1172. doi: 10.11834/jig.211073. [22] WOO S, PARK J, LEE J Y, et al. CBAM: Convolutional block attention module[C]. Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 2018: 3–19. doi: 10.1007/978-3-030-01234-2_1. [23] CHENG Jun. Brain tumor dataset[EB/OL]. https://figshare.com/articles/dataset/brain_tumor_dataset/1512427, 2024. [24] REDDY C K K, REDDY P A, JANAPATI H, et al. A fine-tuned vision transformer based enhanced multi-class brain tumor classification using MRI scan imagery[J]. Frontiers in Oncology, 2024, 14: 1400341. doi: 10.3389/fonc.2024.1400341. [25] ALAM N, ZHU Yutong, SHAO Jiaqi, et al. A novel deep learning framework for brain tumor classification using improved Swin transformer V2[J]. ICCK Transactions on Advanced Computing and Systems, 2025, 1(3): 154–163. doi: 10.62762/tacs.2025.807755. [26] Haque R, Hassan M, Bairagi A K, et al. NeuroNet19: An explainable deep neural network model for the classification of brain tumors using magnetic resonance imaging data[J]. Scientific Reports, 2024, 14(1): 1524. doi: 10.1038/s41598-024-51867-1. [27] KARAGOZ M A, NALBANTOGLU O U, and FOX G C. Residual vision transformer (ResViT) based self-supervised learning model for brain tumor classification[EB/OL]. https://arxiv.org/abs/2411.12874, 2024. [28] WU Kan, ZHANG Jinnian, PENG Houwen, et al. TinyViT: Fast pretraining distillation for small vision transformers[C]. 17th European Conference on Computer Vision, Tel Aviv, Israel, 2022: 68–85. doi: 10.1007/978-3-031-19803-8_5. [29] GRAHAM B, EL-NOUBY A, TOUVRON H, et al. LeViT: A vision transformer in ConvNet's clothing for faster inference[C]. Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 12239–12249. doi: 10.1109/ICCV48922.2021.01204. [30] QIN Danfeng, LEICHNER C, DELAKIS M, et al. MobileNetV4: Universal models for the mobile ecosystem[C]. 18th European Conference on Computer Vision, Milan, Italy, 2024: 78–96. doi: 10.1007/978-3-031-73661-2_5. [31] GÓMEZ-GUZMÁN M A, JIMÉNEZ-BERISTAÍN L, GARCÍA-GUERRERO E E, et al. Classifying brain tumors on magnetic resonance imaging by using convolutional neural networks[J]. Electronics, 2023, 12(4): 955. doi: 10.3390/electronics12040955. [32] DAS S, GHOSH P, and CHAUDHURI A K. Segmentation and classification of specific pattern of Brain tumor using CNN[J]. International Journal of Engineering Technology and Management Sciences, 2023, 7(2): 21–29. doi: 10.46647/ijetms.2023.v07i02.004. [33] RAOUF M H G, FALLAH A, and RASHIDI S. Use of discrete cosine-based stockwell transform in the binary classification of magnetic resonance images of brain tumor[C]. 2022 29th National and 7th International Iranian Conference on Biomedical Engineering (ICBME), Tehran, Iran, Islamic Republic of, 2022: 293–298. doi: 10.1109/ICBME57741.2022.10052875. [34] ELHADIDY M S, ELGOHR A T, EL-GENEEDY M, et al. Comparative analysis for accurate multi-classification of brain tumor based on significant deep learning models[J]. Computers in Biology and Medicine, 2025, 188: 109872. doi: 10.1016/j.compbiomed.2025.109872. [35] DUTTA T K, NAYAK D R, and ZHANG Yudong. ARM-Net: Attention-guided residual multiscale CNN for multiclass brain tumor classification using MR images[J]. Biomedical Signal Processing and Control, 2024, 87: 105421. doi: 10.1016/j.bspc.2023.105421. [36] DUTTA T K, NAYAK D R, and PACHORI R B. GT-Net: Global transformer network for multiclass brain tumor classification using MR images[J]. Biomedical Engineering Letters, 2024, 14(5): 1069–1077. doi: 10.1007/s13534-024-00393-0. [37] GHASSEMI N, SHOEIBI A, and ROUHANI M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images[J]. Biomedical Signal Processing and Control, 2020, 57: 101678. doi: 10.1016/j.bspc.2019.101678. [38] DEEPAK S and AMEER P M. Brain tumor classification using deep CNN features via transfer learning[J]. Computers in Biology and Medicine, 2019, 111: 103345. doi: 10.1016/j.compbiomed.2019.103345. [39] RIZWAN M, SHABBIR A, JAVED A R, et al. Brain tumor and glioma grade classification using Gaussian convolutional neural network[J]. IEEE Access, 2022, 10: 29731–29740. doi: 10.1109/ACCESS.2022.3153108. [40] HAQ A U, LI Jianping, KHAN S, et al. DACBT: Deep learning approach for classification of brain tumors using MRI data in IoT healthcare environment[J]. Scientific Reports, 2022, 12(1): 15331. doi: 10.1038/s41598-022-19465-1. [41] GAB ALLAH A M, SARHAN A M, and ELSHENNAWY N M. Classification of brain MRI tumor images based on deep learning PGGAN augmentation[J]. Diagnostics, 2021, 11(12): 2343. doi: 10.3390/diagnostics11122343. [42] SZEGEDY C, LIU Wei, JIA Yangqing, et al. Going deeper with convolutions[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, USA, 2015: 1–9. doi: 10.1109/CVPR.2015.7298594. [43] DOSOVITSKIY A, BEYER L, KOLESNIKOV A, et al. An image is worth 16x16 words: Transformers for image recognition at scale[C]. Proceedings of the 9th International Conference on Learning Representations, Vienna, Austria, 2020. (查阅网上资料, 未找到本条文献页码信息, 请确认). [44] VAN DER MAATEN L, HINTON G. Visualizing data using t-SNE[J]. Journal of Machine Learning Research, 2008, 9: 2579–2605. -





 下载:
下载:
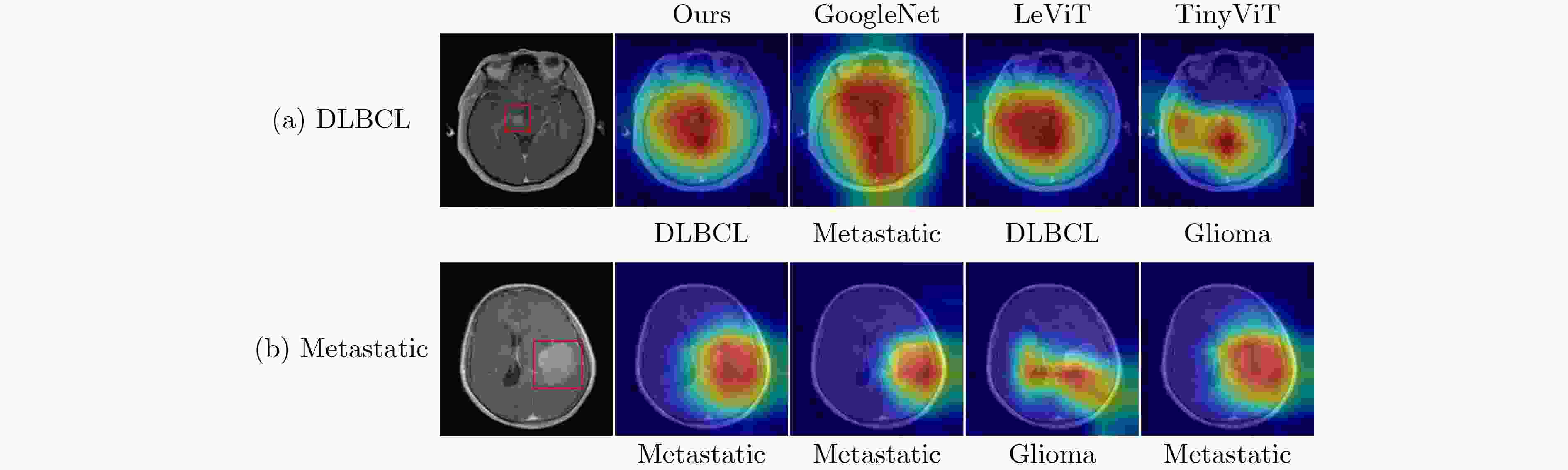
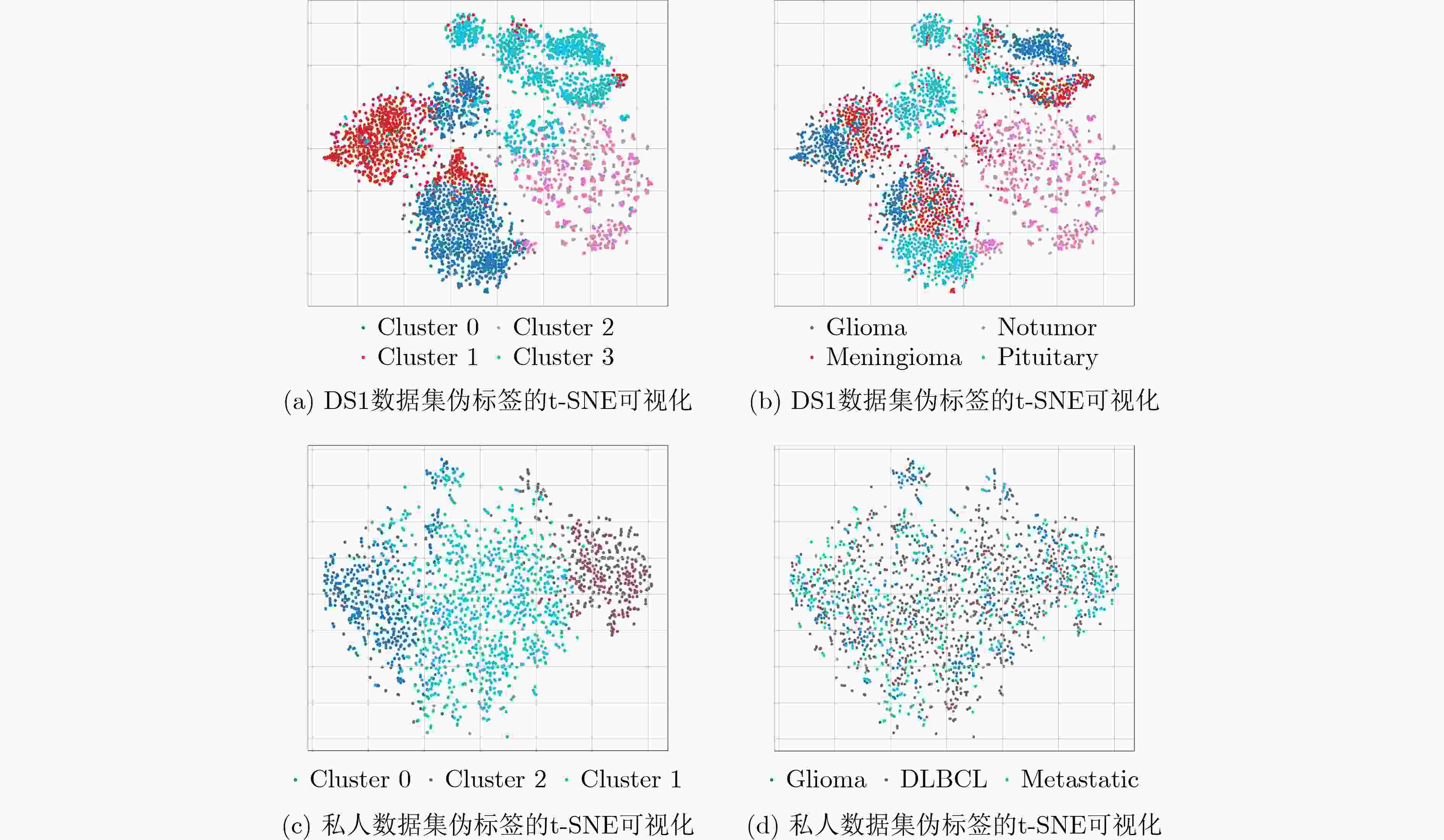
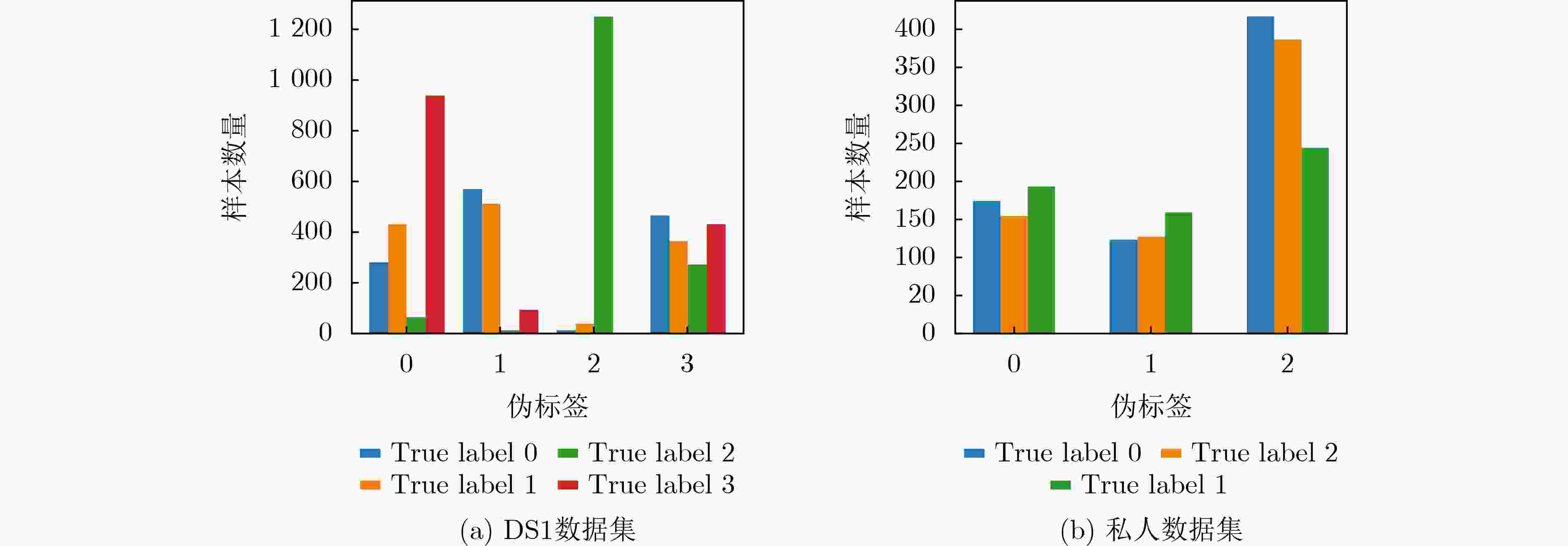


 下载:
下载:
