Privacy-Preserving Federated Weakly-Supervised Learning for Cancer Subtyping on Histopathology Images
-
摘要: 数据驱动的深度学习方法已展现出优越性能,但其成功实施往往依赖于大量细粒度标注训练数据。此外,医疗数据通常呈“数据孤岛”状态,复杂的数据共享过程可能会存在患者隐私泄露的风险。联邦学习 (FL)能够使多个医疗中心在不共享数据的情况下协同训练一个深度学习模型。然而,在计算病理学领域,源自不同医疗中心的病理图像之间普遍存在数据异质性。这些固有的数据异质性可能会显著影响模型性能。针对以上问题,该研究提出一种适用于计算病理学领域千兆像素全切片图像 (WSI)的隐私保护FL方法,该方法结合弱监督的注意力多实例学习 (MIL)与差分隐私技术。具体而言,对于各个参与客户端,使用一种弱监督的多尺度注意力MIL方法,仅需要切片级标签监督本地模型训练,以应对千兆像素病理WSI标注成本高昂的问题。在联邦权重聚合阶段,引入本地化差分隐私技术,进一步降低敏感数据泄露风险;同时采用一种新的联邦自适应重加权策略,旨在克服客户端之间病理图像异质性所带来的挑战。在两种癌症组织学分型任务上评估了所提出FL方法的有效性。实验结果表明,在保障患者数据隐私的前提下,该研究所提出的FL方法相较于本地化模型及其它FL方法,表现出更高的分类准确率;即便与中心化模型相比,其分类性能仍然具备一定竞争力。Abstract:
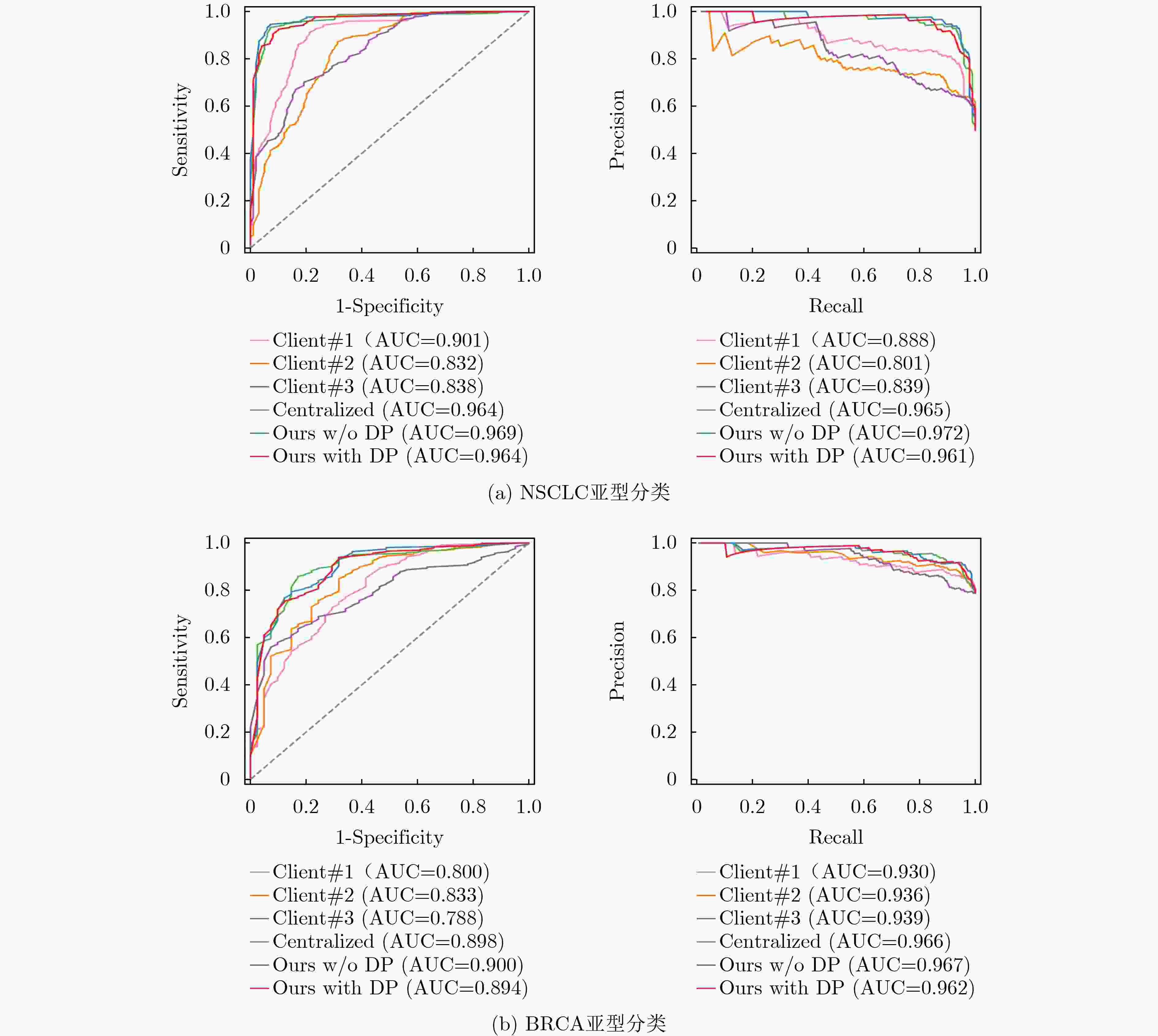
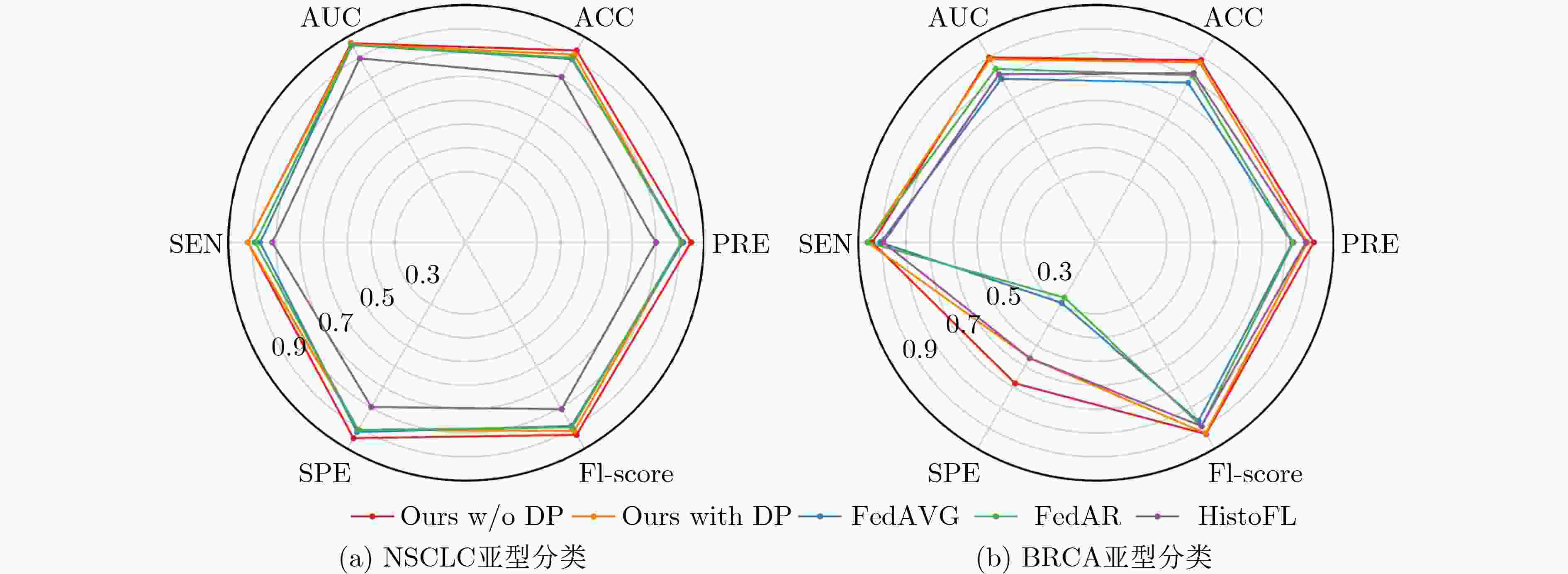
Objective Data-driven deep learning methods have demonstrated superior performance. The development of robust and accurate models often relies on a large amount of training data with fine-grained annotations, which incurs high annotation costs for gigapixel whole slide images (WSI) in histopathology. Typically, healthcare data exists in “data silos”, and the complex data sharing process may raise privacy concerns. Federated Learning (FL) is a promising approach that enables training a global model from data spread across numerous medical centers without exchanging data. However, in traditional FL algorithms, the inherent data heterogeneity across medical centers significantly impacts the performance of the global model. Methods In response to these challenges, this work proposes a privacy-preserving FL method for gigapixel WSIs in computational pathology. The method integrates weakly supervised attention-based multiple instance learning (MIL) with differential privacy techniques. In the context of each client, a multi-scale attention-based MIL method is employed for local training on histopathology WSIs, with only slide-level labels available. This effectively mitigates the high costs of pixel-level annotation for histopathology WSIs via a weakly supervised setting. In the federated model update phase, local differential privacy is used to further mitigate the risk of sensitive data leakage. Specifically, random noise that follows a Gaussian or Laplace distribution is added to the model parameters after local training on each client. Furthermore, a novel federated adaptive reweighting strategy is adopted to overcome challenges posed by the heterogeneity of pathological images across clients. This strategy dynamically balances the contribution of the quantity and quality of local data to each client's weight. Results and Discussions The proposed FL framework is evaluated on two clinical diagnostic tasks: Non-small Cell Lung Cancer (NSCLC) histologic subtyping and Breast Invasive Carcinoma (BRCA) histologic subtyping. As shown in ( Table 1 ,Table 2 , andFig. 4 ), the proposed FL method (Ours with DP and Ours w/o DP) exhibits superior accuracy and generalization when compared with both localized models and other FL methods. Notably, even when compared to the centralized model, its classification performance remains competitive (Fig. 3 ). These results demonstrate that privacy-preserving FL not only serves as a feasible and effective method for multicenter histopathology images, but also may mitigate the performance degradation typically caused by data heterogeneity across centers. By controlling the intensity of added noise within a limited range, the model can also achieve stable classification (Table 3 ). The two key components (i.e., multi-scale representation attention network and federated adaptive reweighting strategy) are proven valuable for consistent performance improvement (Table 4 ). In addition, the proposed FL method maintains stable classification performance across different hyperparameter settings (Table 5 ,Table 6 ). These results further demonstrate that the proposed FL method is robust.Conclusions In conclusion, the proposed FL method tackles two critical issues in multicenter computational pathology: data silos and privacy concerns. Moreover, it can effectively alleviates the performance degradation induced by inter-center data heterogeneity. Given the challenges in balancing model accuracy and privacy protection, future work will explore new methods that preserve privacy while maintaining model performance. -
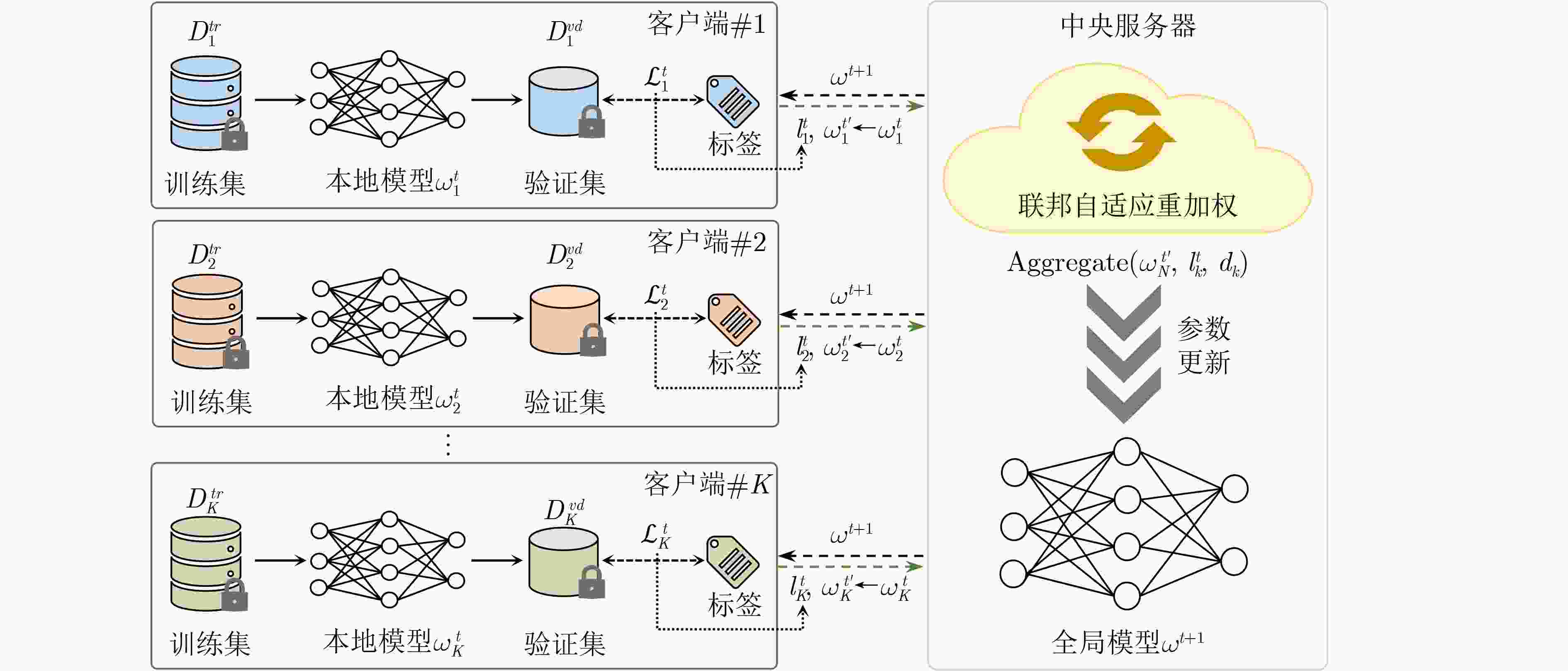
1 多尺度表征注意力网络
输入: 训练集病理WSIs $ {\left\{{I}_{n}\right\}}_{n=1}^{N} $,标签集$ {\left\{{Y}_{n}\right\}}_{n=1}^{N} $,特征提取
器$ f(\cdot ) $,第一个分支中的全连接层$ {h}_{1}(\cdot ) $,第二个分支中的全连
接层$ {h}_{2}(\cdot ) $,注意力模块$ Atten(\cdot ) $输出: 病理WSI的预测类向量$ {P}_{n}^{w} $ 1: $ {\left\{{I}_{ni}\right\}}_{i=1}^{{M}_{b}}\leftarrow {I}_{n} $ //将WSI切割成$ {M}_{b} $张包级图像 2: $ {\gamma }_{ni}\leftarrow 1 $ //初始化WSI中每张包级图像的注意力权重 3: for 每次迭代$ t\in \{1,2,\cdots ,T\} $ do //第一个分支 4: for 每个minibatch do 5: $ {\left\{{I}_{nij}\right\}}_{j=1}^{{M}_{p}}\leftarrow {I}_{ni} $ //将包级图像划分为$ {M}_{p} $张补丁级图像 6: $ {\left\{{Z}_{nijk}^{c}\right\}}_{k=1}^{{M}_{c}}\leftarrow f\left({I}_{nij}\right) $ 7: $ {\left\{{\eta }_{nijk}\right\}}_{k=1}^{{M}_{c}}\leftarrow Atten\left({h}_{1}\left({\left\{{Z}_{nijk}^{c}\right\}}_{k=1}^{{M}_{c}}\right)\right) $ //细胞级
注意力权重8: $ {Z}_{nij}^{p}\leftarrow {\sum }_{k=1}^{{M}_{c}}{\eta }_{nijk}\cdot {Z}_{nijk}^{c} $ 9: $ {\left\{{\delta }_{nij}\right\}}_{j=1}^{{M}_{p}}\leftarrow Atten\left({h}_{1}\left({\left\{{Z}_{nij}^{p}\right\}}_{j=1}^{{M}_{p}}\right)\right) $ //补丁级注
意力权重10: $ {Z}_{ni}^{b}\leftarrow {\sum }_{j=1}^{{M}_{p}}{\delta }_{nij}{\cdot Z}_{nij}^{p} $ 11: end //第二个分支 12: for每个minibatch do 13: $ {\left\{{\gamma }_{ni}\right\}}_{i=1}^{{M}_{b}}\leftarrow Atten\left({h}_{2}\right({\left\{{Z}_{ni}^{b}\right\}}_{i=1}^{{M}_{b}}\left)\right) $ //包级注意力权重 14: $ {Z}_{n}^{w}\leftarrow \displaystyle\sum\nolimits_{i=1}^{{M}_{b}}{\gamma }_{ni}{\cdot Z}_{ni}^{b} $ 15: $ {P}_{n}^{w}\leftarrow h\left({Z}_{n}^{w}\right) $ 16: end 17: end 表 1 与基线方法的对比结果
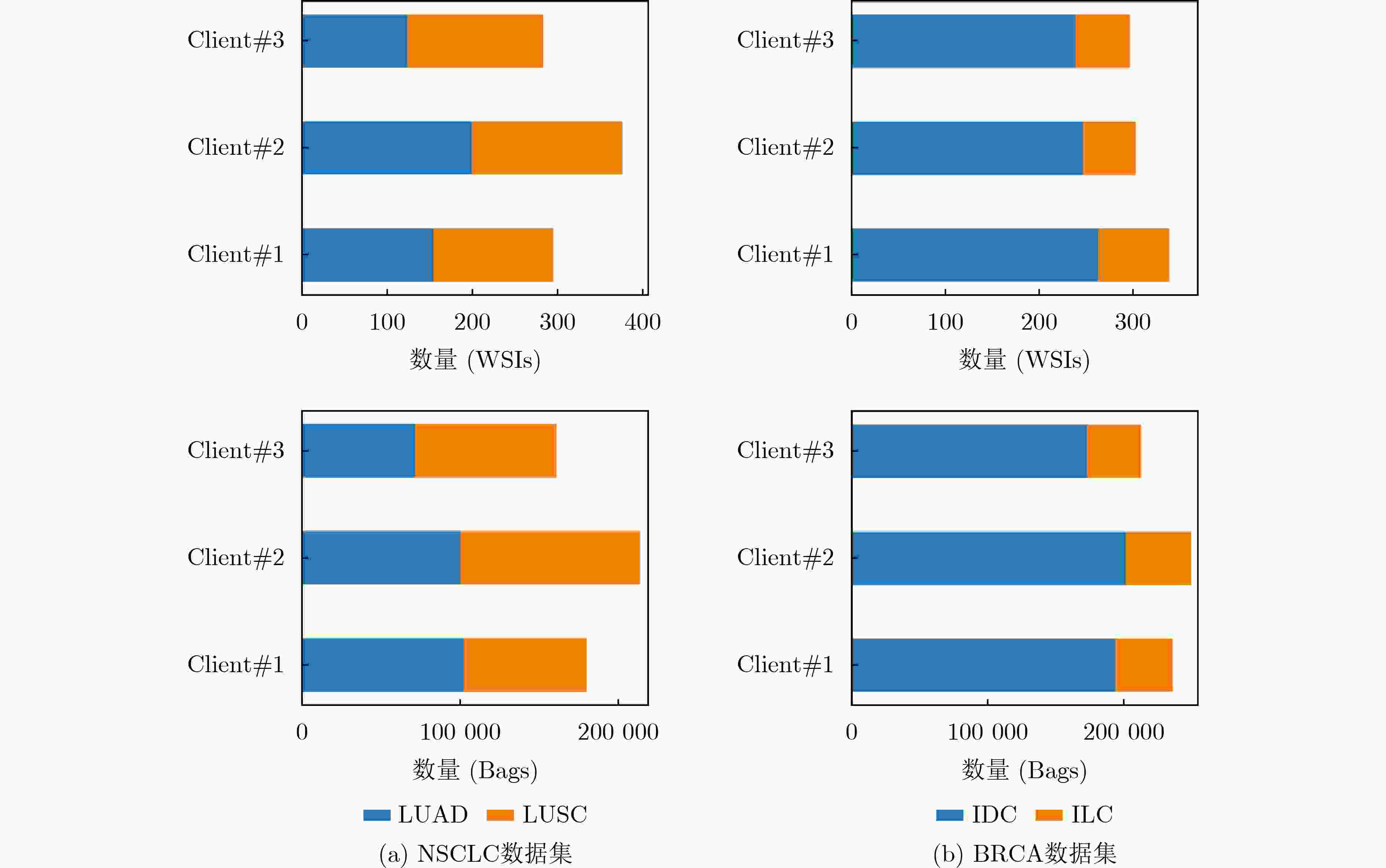
数据集 模型 AUC ACC PRE SEN SPE F1-score P NSCLC Client #1 0.901 0.777 0.837 0.684 0.869 0.753 <0.001 Client #2 0.832 0.751 0.738 0.776 0.727 0.756 0.034 Client #3 0.838 0.716 0.678 0.816 0.616 0.741 <0.001 Centralized 0.964 0.929 0.938 0.918 0.939 0.928 0.003 Ours w/o DP 0.969 0.934 0.947 0.918 0.949 0.933 - Ours with DP 0.964 0.914 0.909 0.918 0.909 0.914 0.023 BRCA Client #1 0.800 0.814 0.877 0.889 0.537 0.883 0.045 Client #2 0.833 0.845 0.855 0.967 0.390 0.908 <0.001 Client #3 0.788 0.763 0.809 0.915 0.195 0.859 <0.001 Centralized 0.898 0.881 0.906 0.948 0.634 0.927 0.124 Ours w/o DP 0.900 0.887 0.917 0.941 0.683 0.929 - Ours with DP 0.892 0.876 0.891 0.961 0.561 0.925 0.702 表 2 与其他FL方法的对比结果
数据集 模型 AUC ACC PRE SEN SPE F1-score P NSCLC FedAVG 0.960 0.893 0.914 0.867 0.919 0.890 <0.001 FedAR 0.961 0.898 0.906 0.888 0.909 0.897 0.006 HistoFL 0.896 0.807 0.800 0.816 0.798 0.808 0.013 Ours w/o DP 0.969 0.934 0.947 0.918 0.949 0.933 - Ours with DP 0.964 0.914 0.909 0.918 0.909 0.914 0.023 BRCA FedAVG 0.797 0.778 0.827 0.908 0.293 0.866 <0.001 FedAR 0.845 0.814 0.831 0.961 0.268 0.891 0.002 HistoFL 0.819 0.825 0.884 0.895 0.561 0.890 <0.001 Ours w/o DP 0.900 0.887 0.917 0.941 0.683 0.929 - Ours with DP 0.892 0.876 0.891 0.961 0.561 0.925 0.702 表 3 添加不同程度噪声对本文方法的分类性能影响
添加噪声
程度NSCLC BRCA AUC ACC F1-score AUC ACC F1-score 高斯噪声 0.1 0.845 0.751 0.717 0.706 0.799 0.887 0.01 0.963 0.914 0.914 0.876 0.876 0.924 0.001 0.964 0.914 0.914 0.892 0.876 0.925 0.0001 0.957 0.909 0.910 0.894 0.892 0.932 拉普拉斯噪声 0.1 0.767 0.726 0.722 0.593 0.789 0.882 0.01 0.971 0.914 0.913 0.897 0.892 0.933 0.001 0.958 0.909 0.909 0.892 0.876 0.925 0.0001 0.963 0.924 0.924 0.872 0.892 0.933 - 0 0.969 0.934 0.933 0.900 0.887 0.929 表 4 NSCLC数据集上,本文方法采用不同基础组件组合的定量结果
基线模型 联邦自适应重加权 多尺度表征注意力 AUC ACC PRE SEN SPE F1-score √ 0.896 0.807 0.800 0.816 0.798 0.808 √ 0.930 0.873 0.892 0.847 0.899 0.869 √ 0.960 0.893 0.914 0.867 0.919 0.890 √ √ 0.969 0.934 0.947 0.918 0.949 0.933 表 5 NSCLC数据集上,本文方法在不同客户端数量下的性能表现
客户端数量 模型 AUC ACC PRE SEN SPE F1-score 3 Localized 0.857 0.748 0.751 0.759 0.737 0.750 HistoFL 0.896 0.807 0.800 0.816 0.798 0.808 Ours 0.969 0.934 0.947 0.918 0.949 0.933 4 Localized 0.747 0.680 0.675 0.693 0.668 0.677 HistoFL 0.855 0.766 0.823 0.670 0.860 0.739 Ours 0.931 0.873 0.860 0.887 0.860 0.873 5 Localized 0.754 0.657 0.653 0.742 0.572 0.671 HistoFL 0.898 0.819 0.796 0.860 0.778 0.827 Ours 0.939 0.899 0.900 0.900 0.899 0.900 表 6 不同超参数(β和λ)设置下,本文方法在NSCLC数据集上的性能表现
β λ AUC ACC PRE SEN SPE F1-score 0.5 1 0.945 0.883 0.879 0.888 0.879 0.883 5 0.958 0.904 0.899 0.908 0.899 0.904 10 0.963 0.898 0.898 0.898 0.899 0.898 1 1 0.957 0.898 0.906 0.888 0.909 0.897 5 0.954 0.914 0.918 0.908 0.919 0.913 10 0.969 0.934 0.947 0.918 0.949 0.933 1.5 1 0.928 0.878 0.894 0.857 0.899 0.875 5 0.946 0.904 0.876 0.939 0.869 0.906 10 0.948 0.888 0.913 0.857 0.919 0.884 -
[1] BRAY F, LAVERSANNE M, SUNG H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2024, 74(3): 229–263. doi: 10.3322/caac.21834. [2] HAN Bingfeng, ZHENG Rongshou, ZENG Hongmei, et al. Cancer incidence and mortality in China, 2022[J]. Journal of the National Cancer Center, 2024, 4(1): 47–53. doi: 10.1016/j.jncc.2024.01.006. [3] DENTRO S C, LESHCHINER I, HAASE K, et al. Characterizing genetic intra-tumor heterogeneity across 2, 658 human cancer genomes[J]. Cell, 2021, 184(8): 2239–2254. e39. doi: 10.1016/j.cell.2021.03.009. [4] WANG Yibei, SAFI M, HIRSCH F R, et al. Immunotherapy for advanced-stage squamous cell lung cancer: The state of the art and outstanding questions[J]. Nature Reviews Clinical Oncology, 2025, 22(3): 200–214. doi: 10.1038/s41571-024-00979-8. [5] GONG Tingting, GUO Shuang, LIU Fanghua, et al. Proteomic characterization of epithelial ovarian cancer delineates molecular signatures and therapeutic targets in distinct histological subtypes[J]. Nature Communications, 2023, 14(1): 7802. doi: 10.1038/s41467-023-43282-3. [6] NASRAZADANI A, LI Yujia, FANG Yusi, et al. Mixed invasive ductal lobular carcinoma is clinically and pathologically more similar to invasive lobular than ductal carcinoma[J]. British Journal of Cancer, 2023, 128(6): 1030–1039. doi: 10.1038/s41416-022-02131-8. [7] ELMORE J. Abstract SY01–03: The gold standard cancer diagnosis: Studies of physician variability, interpretive behavior, and the impact of AI[J]. Cancer Research, 2021, 81(S13): SY01–03. doi: 10.1158/1538-7445.AM2021-SY01-03. [8] MADABHUSHI A and LEE G. Image analysis and machine learning in digital pathology: Challenges and opportunities[J]. Medical Image Analysis, 2016, 33: 170–175. doi: 10.1016/j.media.2016.06.037. [9] LI Bin, KEIKHOSRAVI A, LOEFFLER A G, et al. Single image super-resolution for whole slide image using convolutional neural networks and self-supervised color normalization[J]. Medical Image Analysis, 2021, 68: 101938. doi: 10.1016/j.media.2020.101938. [10] BULTEN W, PINCKAERS H, VAN BOVEN H, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: A diagnostic study[J]. The Lancet Oncology, 2020, 21(2): 233–241. doi: 10.1016/S1470-2045(19)30739-9. [11] SRINIDHI C L, CIGA O, and MARTEL A L. Deep neural network models for computational histopathology: A survey[J]. Medical Image Analysis, 2021, 67: 101813. doi: 10.1016/j.media.2020.101813. [12] DIETTERICH T G, LATHROP R H, and LOZANO-PÉREZ T. Solving the multiple instance problem with axis-parallel rectangles[J]. Artificial Intelligence, 1997, 89(1/2): 31–71. doi: 10.1016/S0004-3702(96)00034-3. [13] CARBONNEAU M A, CHEPLYGINA V, GRANGER E, et al. Multiple instance learning: A survey of problem characteristics and applications[J]. Pattern Recognition, 2018, 77: 329–353. doi: 10.1016/j.patcog.2017.10.009. [14] LU M Y, WILLIAMSON D F K, CHEN T Y, et al. Data-efficient and weakly supervised computational pathology on whole-slide images[J]. Nature Biomedical Engineering, 2021, 5(6): 555–570. doi: 10.1038/s41551-020-00682-w. [15] BONTEMPO G, BOLELLI F, PORRELLO A, et al. A graph-based multi-scale approach with knowledge distillation for WSI classification[J]. IEEE Transactions on Medical Imaging, 2024, 43(4): 1412–1421. doi: 10.1109/TMI.2023.3337549. [16] DENG Jia, DONG Wei, SOCHER R, et al. ImageNet: A large-scale hierarchical image database[C]. 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, USA, 2009: 248–255. doi: 10.1109/CVPR.2009.5206848. [17] MARELLI L and TESTA G. Scrutinizing the EU general data protection regulation[J]. Science, 2018, 360(6388): 496–498. doi: 10.1126/science.aar5419. [18] MARKS M and HAUPT C E. AI chatbots, health privacy, and challenges to HIPAA compliance[J]. JAMA, 2023, 330(4): 309–310. doi: 10.1001/jama.2023.9458. [19] MCMAHAN B, MOORE E, RAMAGE D, et al. Communication-efficient learning of deep networks from decentralized data[C]. The 20th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, USA, 2017: 1273–1282. [20] KARARGYRIS A, UMETON R, SHELLER M J, et al. Federated benchmarking of medical artificial intelligence with MedPerf[J]. Nature Machine Intelligence, 2023, 5(7): 799–810. doi: 10.1038/s42256-023-00652-2. [21] DU TERRAIL J O, LEOPOLD A, JOLY C, et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer[J]. Nature Medicine, 2023, 29(1): 135–146. doi: 10.1038/s41591-022-02155-w. [22] ZHANG Yuanming, LI Zheng, HAN Xiangmin, et al. Pseudo-data based self-supervised federated learning for classification of histopathological images[J]. IEEE Transactions on Medical Imaging, 2024, 43(3): 902–915. doi: 10.1109/TMI.2023.3323540. [23] RODRÍGUEZ-BARROSO N, JIMÉNEZ-LÓPEZ D, LUZÓN M V, et al. Survey on federated learning threats: Concepts, taxonomy on attacks and defences, experimental study and challenges[J]. Information Fusion, 2023, 90: 148–173. doi: 10.1016/j.inffus.2022.09.011. [24] ZHANG Yuheng, JIA Ruoxi, PEI Hengzhi, et al. The secret revealer: Generative model-inversion attacks against deep neural networks[C]. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, USA, 2020: 250–258. doi: 10.1109/CVPR42600.2020.00033. [25] GEIPING J, BAUERMEISTER H, DRÖGE H, et al. Inverting gradients-how easy is it to break privacy in federated learning?[C]. The 34th International Conference on Neural Information Processing Systems, Vancouver, Canada, 2020: 1421. [26] WANG Zhibo, SONG Mengkai, ZHANG Zhifei, et al. Beyond inferring class representatives: User-level privacy leakage from federated learning[C]. IEEE INFOCOM 2019-IEEE Conference on Computer Communications, Paris, France, 2019: 2512–2520. doi: 10.1109/INFOCOM.2019.8737416. [27] DONG Jinshuo, ROTH A, and SU Weijie. Gaussian differential privacy[J]. Journal of the Royal Statistical Society Series B: Statistical Methodology, 2022, 84(1): 3–37. doi: 10.1111/rssb.12454. [28] KAISSIS G A, MAKOWSKI M R, RÜCKERT D, et al. Secure, privacy-preserving and federated machine learning in medical imaging[J]. Nature Machine Intelligence, 2020, 2(6): 305–311. doi: 10.1038/s42256-020-0186-1. [29] WANG Xiaoding, HU Jia, LIN Hui, et al. Federated learning-empowered disease diagnosis mechanism in the internet of medical things: From the privacy-preservation perspective[J]. IEEE Transactions on Industrial Informatics, 2023, 19(7): 7905–7913. doi: 10.1109/TII.2022.3210597. [30] XIANG Hangchen, SHEN Junyi, YAN Qingguo, et al. Multi-scale representation attention based deep multiple instance learning for gigapixel whole slide image analysis[J]. Medical Image Analysis, 2023, 89: 102890. doi: 10.1016/j.media.2023.102890. [31] CHIDAMBARANATHAN M, SHARMA U, NAIDU C M, et al. A new approach for recognition of implant in knee by template matching[J]. Indian Journal of Science and Technology, 2016, 9(37): 1–5. doi: 10.17485/ijst/2016/v9i37/102081. [32] SHI Xiaoshuang, XING Fuyong, XU Kaidi, et al. Loss-based attention for interpreting image-level prediction of convolutional neural networks[J]. IEEE Transactions on Image Processing, 2021, 30: 1662–1675. doi: 10.1109/TIP.2020.3046875. [33] GUO Shengnan, WANG Xibin, LONG Shigong, et al. A federated learning scheme meets dynamic differential privacy[J]. CAAI Transactions on Intelligence Technology, 2023, 8(3): 1087–1100. doi: 10.1049/cit2.12187. [34] ZHENG Yifeng, LAI Shangqi, LIU Yi, et al. Aggregation service for federated learning: An efficient, secure, and more resilient realization[J]. IEEE Transactions on Dependable and Secure Computing, 2023, 20(2): 988–1001. doi: 10.1109/TDSC.2022.3146448. [35] WANG Bo, LI Hongtao, GUO Yina, et al. PPFLHE: A privacy-preserving federated learning scheme with homomorphic encryption for healthcare data[J]. Applied Soft Computing, 2023, 146: 110677. doi: 10.1016/j.asoc.2023.110677. [36] LI Xiaoxiao, GU Yufeng, DVORNEK N, et al. Multi-site fMRI analysis using privacy-preserving federated learning and domain adaptation: ABIDE results[J]. Medical Image Analysis, 2020, 65: 101765. doi: 10.1016/j.media.2020.101765. [37] LU M Y, CHEN R J, KONG Dehan, et al. Federated learning for computational pathology on gigapixel whole slide images[J]. Medical Image Analysis, 2022, 76: 102298. doi: 10.1016/j.media.2021.102298. [38] MACENKO M, NIETHAMMER M, MARRON J S, et al. A method for normalizing histology slides for quantitative analysis[C]. 2019 IEEE International Symposium on Biomedical Imaging, Boston, USA, 2009: 1107–1110. doi: 10.1109/ISBI.2009.5193250. [39] MA Benteng, FENG Yu, CHE Geng, et al. Federated adaptive reweighting for medical image classification[J]. Pattern Recognition, 2023, 144: 109880. doi: 10.1016/j.patcog.2023.109880. [40] ILSE M, TOMCZAK J M, and WELLING M. Attention-based deep multiple instance learning[C]. The 35th International Conference on Machine Learning, Stockholmsmässan, Sweden, 2018: 2132–2141. -





 下载:
下载:


 下载:
下载:
