Federated Semi-Supervised Image Segmentation with Dynamic Client Selection
-
摘要: 多中心协同验证是临床研究的必然趋势,但患者隐私保护、跨机构数据分布异质性以及精确标注稀缺,使传统集中式医学图像分割方法难以直接应用。为此,本文提出一种融合动态客户端管理机制的联邦半监督医学图像分割框架,利用客户端性能驱动的加权聚合和教师–学生知识蒸馏,在保护隐私的前提下充分挖掘无标签客户端价值;并设计多尺度特征融合UNet (Multi-scale Feature Fusion UNet, MFF-UNet)作为分割骨干,以增强多中心异构影像的特征表征能力,实现对前列腺区域的精准分割。基于来自6家医疗机构的T2加权前列腺MRI数据的实验表明,本方法在有标签和无标签客户端上分别获得
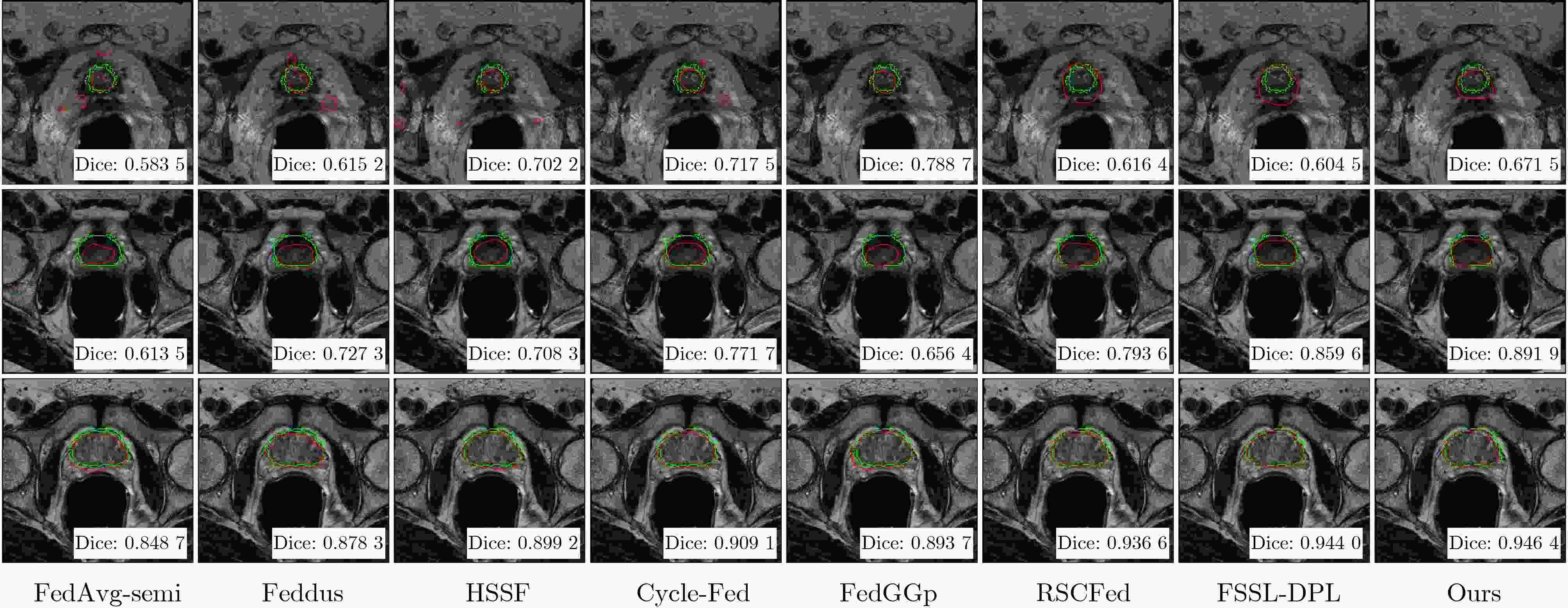
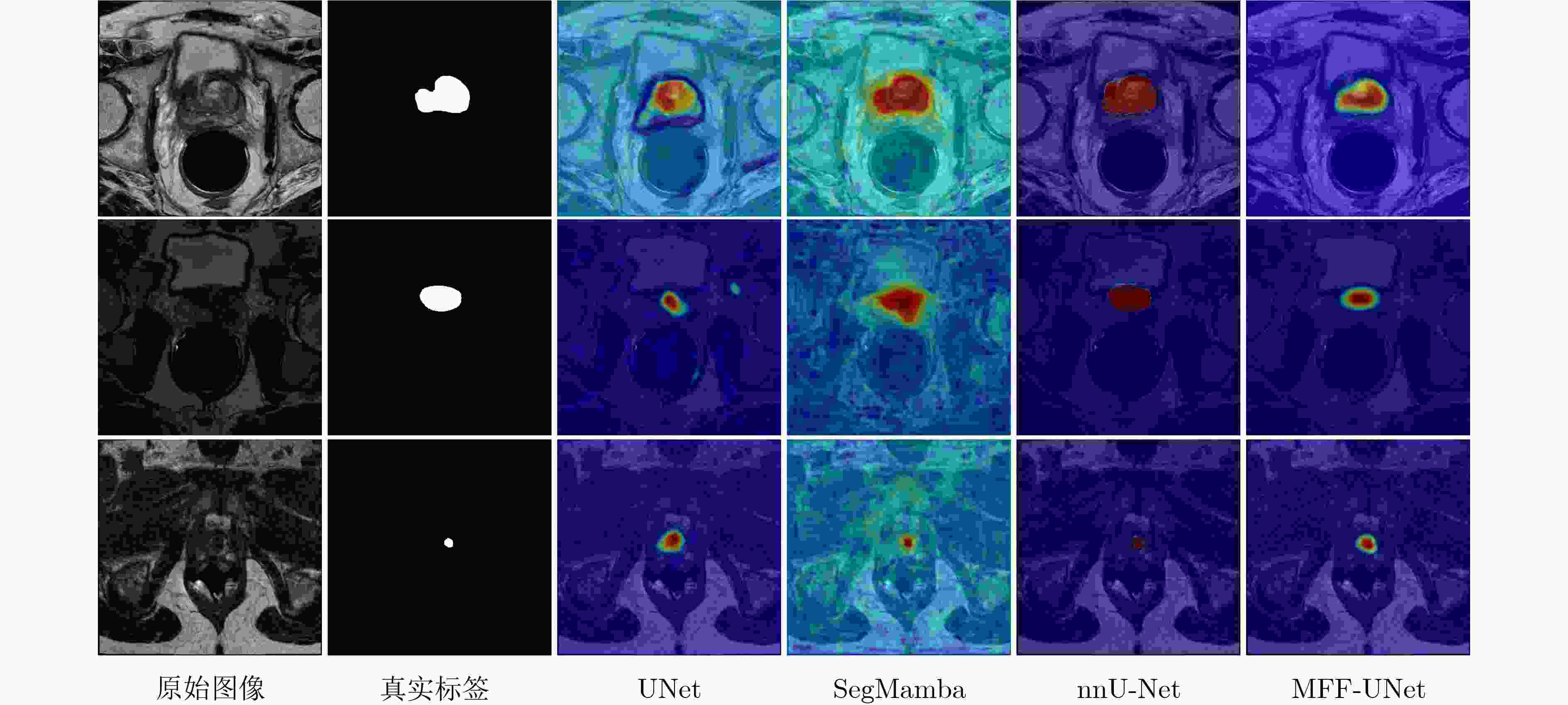
0.8405 /0.7868 的Dice系数和8.04/8.67的HD95,均优于多种现有联邦半监督医学图像分割方法。Abstract:Objective Multicenter validation is an inevitable trend in clinical research, yet strict privacy regulations, heterogeneous cross-institutional data distributions and scarce pixel-level annotations limit the applicability of conventional centralized medical image segmentation models. This study aims to develop a federated semi-supervised framework that jointly exploits labelled and unlabeled prostate MRI data from multiple hospitals, explicitly considering dynamic client participation and non-independent and identically distributed (Non-IID) data, so as to improve segmentation accuracy and robustness under real-world constraints. Methods A cross-silo federated semi-supervised learning paradigm is adopted, in which clients with pixel-wise annotations act as labeled clients and those without annotations act as unlabeled clients. Each client maintains a local student network for prostate segmentation. On unlabeled clients, a teacher network with the same architecture is updated by the exponential moving average of student parameters and generates perturbed pseudo-labels to supervise the student through a hybrid consistency loss that combines Dice and binary cross-entropy terms. To mitigate the negative influence of heterogeneous and low-quality updates, a performance-driven dynamic client selection and aggregation strategy is introduced. At each communication round, clients are evaluated on their local validation sets, and only those whose Dice scores exceed a threshold are retained; then a Top-K subset is aggregated with normalized contribution weights derived from validation Dice, with bounds to avoid gradient vanishing and single-client dominance. For unlabeled clients, a penalty factor is applied to down-weight unreliable pseudo-labeled updates. As the segmentation backbone, a Multi-scale Feature Fusion U-Net (MFF-UNet) is constructed. Starting from a standard encoder–decoder U-Net, an FPN-like pyramid is inserted into the encoder, where multi-level feature maps are channel-aligned by 1×1 convolutions, fused in a top–down pathway by upsampling and element-wise addition, and refined using 3×3 convolutions. The decoder progressively upsamples these fused features and combines them with encoder features via skip connections, enabling joint modelling of global semantics and fine-grained boundaries. The framework is evaluated on T2-weighted prostate MRI from six centers: three labeled clients and three unlabeled clients. All 3D volumes are resampled, sliced into 2D axial images, resized and augmented. Dice coefficient and 95th percentile Hausdorff distance (HD95) are used as evaluation metrics. Results and Discussions On the six-center dataset, the proposed method achieves average Dice scores of 0.8405 on labeled clients and0.7868 on unlabeled clients, with corresponding HD95 values of 8.04 and 8.67 pixels, respectively. These results are consistently superior to or on par with several representative federated semi-supervised or mixed-supervision methods, and the improvements are most pronounced on distribution-shifted unlabeled centers. Qualitative visualization shows that the proposed method produces more complete and smoother prostate contours with fewer false positives in challenging low-contrast or small-volume cases, compared with the baselines. Attention heatmaps extracted from the final decoder layer demonstrate that U-Net suffers from attention drift, SegMamba displays diffuse responses and nnU-Net exhibits weak activations for small lesions, whereas MFF-UNet focuses more precisely on the prostate region with stable high responses, indicating enhanced discriminative capability and interpretability.Conclusions A federated semi-supervised prostate MRI segmentation framework that integrates teacher–student consistency learning, multi-scale feature fusion and performance-driven dynamic client selection is presented. The method preserves patient privacy by keeping data local, alleviates annotation scarcity by exploiting unlabeled clients and explicitly addresses client heterogeneity through reliability-aware aggregation. Experiments on a six-center dataset demonstrate that the proposed framework achieves competitive or superior overlap and boundary accuracy compared with state-of-the-art federated semi-supervised methods, particularly on distribution-shifted unlabeled centers. The framework is model-agnostic and can be extended to other organs, imaging modalities and cross-institutional segmentation tasks under stringent privacy and regulatory constraints. -
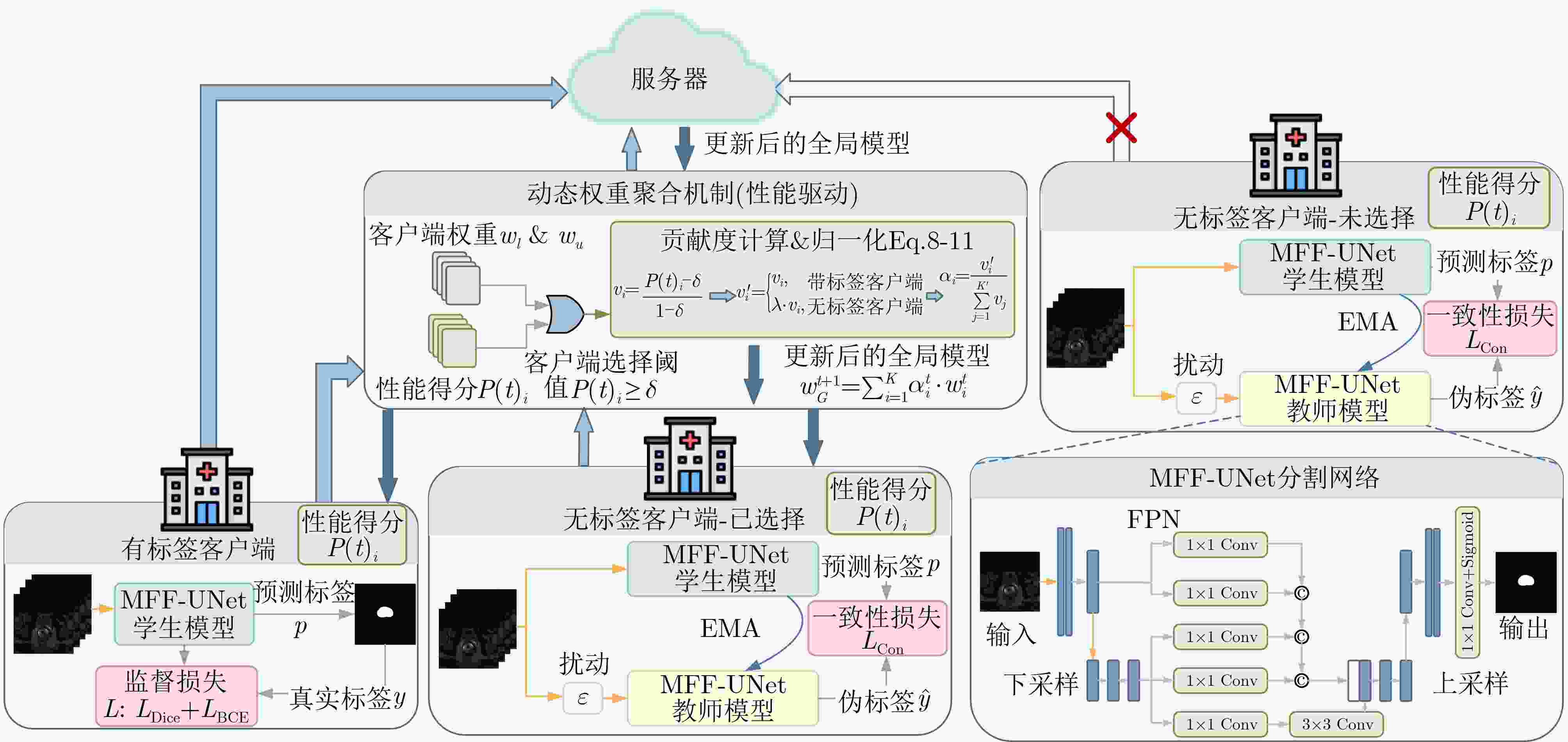
1 引入客户端选择动态调整的联邦半监督学习算法
输入:客户端数据集$ {D}_{i}(i\in [1,1,\cdots,K]) $,带标签客户端模型
$ {w}_{l} $,无标签客户端模型$ {w}_{u} $,总通信轮次$ T(t\in [1,2,\cdots ,T]) $,预
热通信轮次$ {T}_{\text{warmup}} < T $输出:全局模型参数$ {w}_{G} $ 1: 初始化全局模型$ {w}_{G} $; 2: 分发到每个客户端$ {w}_{l},{w}_{u}\leftarrow {w}_{G} $; 3: 将客户端模型传给教师模型$ {w}_{T}\leftarrow {w}_{u,l} $; 4: for $ t\in [1,2,\cdots,T] $: 5: if $ t=={T}_{\text{warmup}} $:加入无标签客户端; 6: 客户端本地模型更新: 7: for 参与的客户端同步训练: 8: $ p\leftarrow \text{sigmoid}({w}_{S}(x)) $ 9: if标签存在或$ t < {T}_{\text{warmup}} $: 10: $ {L}_{\text{Con}}=\alpha \cdot {L}_{\text{BCE}}(p,y)+(1-\alpha )\cdot {L}_{\text{Dice}}(p,y) $ 11: else $ \hat{y}\leftarrow \text{sigmoid}({w}_{T}(x+\varepsilon )) $; $ L=\alpha \cdot {L}_{\text{BCE}}(p,\hat{y})+(1-\alpha )\cdot {L}_{\text{Dice}}(p,\hat{y}) $ 12: 更新教师模型$ w_{T}^{t+1}\leftarrow \tau w_{G}^{t}+\left(1-\tau \right)w_{T}^{t} $ 13: 在每个客户端验证计算得到分数$ P{(t)}_{i} $ 14: 动态权重聚合 15: for 每个客户端: 16: if $ P{(t)}_{i} < \delta $:$ {v}_{i}\leftarrow 0 $ else $ {v}_{i}\leftarrow \max ({v}_{\min },\min ((P{(t)}_{i}-\delta )/(1-\delta ),{v}_{\max })) $ 17: if 无标签客户端:$ {{{v}^{\prime}}}_{i}\leftarrow \lambda \cdot {v}_{i} $ 18: 归一化权重$ {\alpha }_{i}\leftarrow \frac{{{{v}^{\prime}}}_{i}}{\displaystyle\sum\limits_{j=1}^{{{K}^{\prime}}}{v}_{j}},{K}^{\prime}=\{j|{v}_{j} > 0\} $ 19: $ w_{G}^{t+1}=\displaystyle\sum\limits_{i=1}^{K}\alpha _{i}^{t}\cdot w_{i}^{t} $ 20: end 表 1 不同联邦半监督模型在多客户端上的Dice系数比较
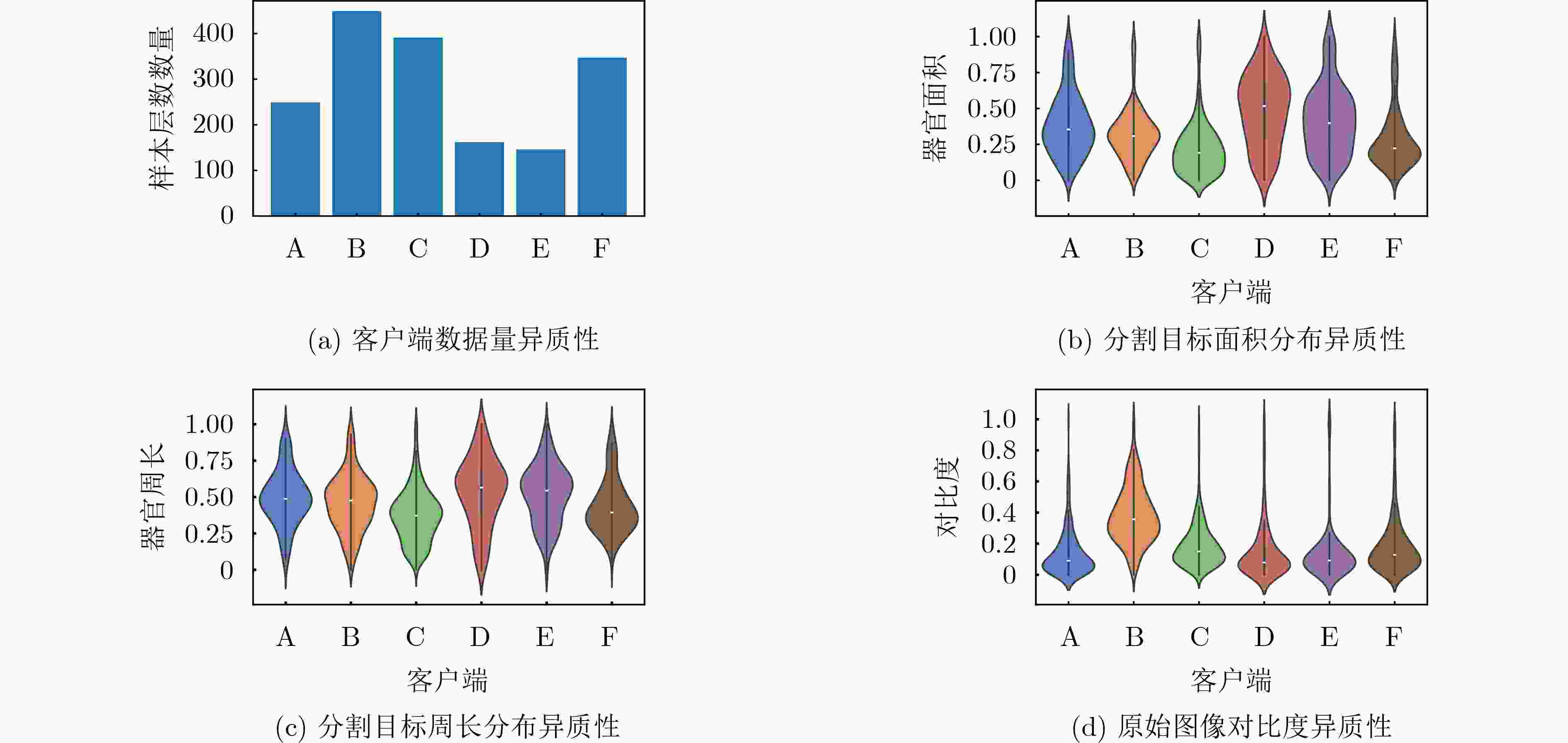
方法 Label Unlabel AvgHD95 客户端 A B C D E F All Label Unlabel FedAvg-semi[33] 0.8009 0.8344 0.8249 0.5998 0.7178 0.6936 0.7452 0.8201 0.6704 Feddus[19] 0.8082 0.8497 0.8179 0.6350 0.7430 0.7428 0.7661 0.8253 0.7069 HSSF[35] 0.7916 0.8148 0.7674 0.7595 0.7455 0.7643 0.7739 0.7913 0.7564 Cycle-Fed[36] 0.7734 0.8640 0.7906 0.7212 0.8128 0.7595 0.7869 0.8093 0.7645 FedGGp[37] 0.7865 0.8485 0.8256 0.7633 0.7902 0.7968 0.8018 0.8202 0.7834 RSCFed[34] 0.7999 0.8126 0.8159 0.6960 0.8291 0.8076 0.7935 0.8095 0.7776 FSSL-DPL[20] 0.8055 0.8462 0.8303 0.7007 0.7987 0.8202 0.8003 0.8273 0.7732 Ours 0.8377 0.8700 0.8138 0.7788 0.8280 0.7537 0.8137 0.8405 0.7868 表 2 不同联邦半监督模型的HD95距离比较(pixel)
方法 Label Unlabel AvgHD95 客户端 A B C D E F All Label Unlabel FedAvg-semi[33] 11.49 14.25 7.93 21.05 11.85 23.23 14.97 11.22 18.71 Feddus[19] 10.70 9.93 9.39 14.02 11.11 15.28 11.74 10.00 13.47 HSSF[35] 12.08 17.17 14.08 17.00 11.96 13.08 14.23 14.44 14.01 Cycle-Fed[36] 13.06 11.00 9.89 19.85 7.50 16.06 12.89 11.31 14.47 FedGGp[37] 10.64 10.98 7.29 17.09 16.17 13.22 12.57 9.64 15.49 RSCFed[34] 13.23 11.10 7.54 11.21 6.92 8.54 9.76 10.62 8.89 FSSL-DPL[20] 10.23 6.87 7.29 10.96 7.38 7.49 8.37 8.13 8.61 Ours 9.30 7.27 7.55 8.23 7.19 10.58 8.35 8.04 8.67 表 3 不同模块配置下的前列腺MRI分割性能对比(Dice)
w/o
动态聚合w/o
MFF-UnetLabel Unlabel A B C D E F 0.8082 0.8497 0.8179 0.6350 0.7430 0.7428 √ 0.8226 0.8242 0.7787 0.6900 0.7855 0.6147 √ 0.8193 0.8516 0.8218 0.6406 0.7612 0.7365 √ √ 0.8377 0.8700 0.8138 0.7788 0.8280 0.7537 -
[1] CORNFORD P, VAN DEN BERGH R C N, BRIERS E, et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG guidelines on prostate cancer—2024 update. Part I: Screening, diagnosis, and local treatment with curative intent[J]. European Urology, 2024, 86(2): 148–163. doi: 10.1016/j.eururo.2024.03.027. [2] 孙军梅, 葛青青, 李秀梅, 等. 一种具有边缘增强特点的医学图像分割网络[J]. 电子与信息学报, 2022, 44(5): 1643–1652. doi: 10.11999/JEIT210784.SUN Junmei, GE Qingqing, LI Xiumei, et al. A medical image segmentation network with boundary enhancement[J]. Journal of Electronics & Information Technology, 2022, 44(5): 1643–1652. doi: 10.11999/JEIT210784. [3] AZAD R, AGHDAM E K, RAULAND A, et al. Medical image segmentation review: The success of U-Net[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2024, 46(12): 10076–10095. doi: 10.1109/TPAMI.2024.3435571. [4] JIANG Yangfan, LUO Xinjian, WU Yuncheng, et al. On data distribution leakage in cross-silo federated learning[J]. IEEE Transactions on Knowledge and Data Engineering, 2024, 36(7): 3312–3328. doi: 10.1109/TKDE.2023.3349323. [5] 肖雄, 唐卓, 肖斌, 等. 联邦学习的隐私保护与安全防御研究综述[J]. 计算机学报, 2023, 46(5): 1019–1044. doi: 10.11897/SP.J.1016.2023.01019.XIAO Xiong, TANG Zhuo, XIAO Bin, et al. A survey on privacy and security issues in federated learning[J]. Chinese Journal of Computers, 2023, 46(5): 1019–1044. doi: 10.11897/SP.J.1016.2023.01019. [6] YANG Xiangli, SONG Zixing, KING I, et al. A survey on deep semi-supervised learning[J]. IEEE Transactions on Knowledge and Data Engineering, 2023, 35(9): 8934–8954. doi: 10.1109/TKDE.2022.3220219. [7] MA Yuxi, WANG Jiacheng, YANG Jing, et al. Model-heterogeneous semi-supervised federated learning for medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2024, 43(5): 1804–1815. doi: 10.1109/TMI.2023.3348982. [8] YANG Dong, XU Ziyue, LI Wenqi, et al. Federated semi-supervised learning for COVID region segmentation in chest CT using multi-national data from China, Italy, Japan[J]. Medical Image Analysis, 2021, 70: 101992. doi: 10.1016/j.media.2021.101992. [9] WU Huisi, ZHANG Baiming, CHEN Cheng, et al. Federated semi-supervised medical image segmentation via prototype-based pseudo-labeling and contrastive learning[J]. IEEE Transactions on Medical Imaging, 2023, 43(2): 649–661. doi: 10.1109/TMI.2023.3314430. [10] LUO Xiangde, WANG Guotai, LIAO Wenjun, et al. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency[J]. Medical Image Analysis, 2022, 80: 102517. doi: 10.1016/j.media.2022.102517. [11] SU Jiawei, LUO Zhiming, LIAN Sheng, et al. Mutual learning with reliable pseudo label for semi-supervised medical image segmentation[J]. Medical Image Analysis, 2024, 94: 103111. doi: 10.1016/j.media.2024.103111. [12] ABUDUWEILI A, LI Xingjian, SHI H, et al. Adaptive consistency regularization for semi-supervised transfer learning[C]. Proceedings of 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, USA, 2021: 6919–6928. doi: 10.1109/CVPR46437.2021.00685. [13] SHELLER M J, EDWARDS B, REINA G A, et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data[J]. Scientific Reports, 2020, 10(1): 12598. doi: 10.1038/s41598-020-69250-1. [14] LIU Quande, DOU Qi, YU Lequan, et al. MS-Net: Multi-site network for improving prostate segmentation with heterogeneous MRI data[J]. IEEE Transactions on Medical Imaging, 2020, 39(9): 2713–2724. doi: 10.1109/TMI.2020.2974574. [15] XU An, LI Wenqi, GUO Pengfei, et al. Closing the generalization gap of cross-silo federated medical image segmentation[C]. Proceedings of 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 20834–20843. doi: 10.1109/CVPR52688.2022.02020. [16] JIANG Meirui, YANG Hongzheng, CHENG Chen, et al. IOP-FL: Inside-outside personalization for federated medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2023, 42(7): 2106–2117. doi: 10.1109/TMI.2023.3263072. [17] LIU Quande, YANG Hongzheng, DOU Qi, et al. Federated semi-supervised medical image classification via inter-client relation matching[C]. Proceedings of the 24th International Conference on Medical Image Computing and Computer Assisted Intervention, Strasbourg, France, 2021: 325–335. doi: 10.1007/978-3-030-87199-4_31. [18] WICAKSANA J, YAN Zengqiang, ZHANG Dong, et al. FedMix: Mixed supervised federated learning for medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2023, 42(7): 1955–1968. doi: 10.1109/TMI.2022.3233405. [19] WANG Dan, HAN Chu, ZHANG Zhen, et al. FedDUS: Lung tumor segmentation on CT images through federated semi-supervised with dynamic update strategy[J]. Computer Methods and Programs in Biomedicine, 2024, 249: 108141. doi: 10.1016/j.cmpb.2024.108141. [20] QIU Liang, CHENG Jierong, GAO Huxin, et al. Federated semi-supervised learning for medical image segmentation via pseudo-label denoising[J]. IEEE Journal of Biomedical and Health Informatics, 2023, 27(10): 4672–4683. doi: 10.1109/JBHI.2023.3274498. [21] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [22] ZHOU Zongwei, RAHMAN SIDDIQUEE M M, TAJBAKHSH N, et al. UNet++: A nested U-Net architecture for medical image segmentation[C]. Proceedings of the 4th International Workshop on Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Granada, Spain, 2018: 3–11. doi: 10.1007/978-3-030-00889-5_1. [23] HUANG Huimin, LIN Lanfen, TONG Ruofeng, et al. UNet 3+: A full-scale connected UNet for medical image segmentation[C]. Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing, Barcelona, Spain, 2020: 1055–1059. doi: 10.1109/ICASSP40776.2020.9053405. [24] LIN T Y, DOLLÁR P, GIRSHICK R, et al. Feature pyramid networks for object detection[C]. Proceedings of 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 936–944. doi: 10.1109/CVPR.2017.106. [25] CHEN Shaolong, QIU Changzhen, YANG Weiping, et al. Combining edge guidance and feature pyramid for medical image segmentation[J]. Biomedical Signal Processing and Control, 2022, 78: 103960. doi: 10.1016/j.bspc.2022.103960. [26] OKTAY O, SCHLEMPER J, LE FOLGOC L, et al. Attention U-Net: Learning where to look for the pancreas[EB/OL]. https://arxiv.org/abs/1804.03999.pdf, 2018. [27] FENG Shuanglang, ZHAO Heming, SHI Fei, et al. CPFNet: Context pyramid fusion network for medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2020, 39(10): 3008–3018. doi: 10.1109/TMI.2020.2983721. [28] GU Zaiwang, CHENG Jun, FU Huazhu, et al. CE-Net: Context encoder network for 2D medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2019, 38(10): 2281–2292. doi: 10.1109/TMI.2019.2903562. [29] DENG Yongheng, LYU Feng, REN Ju, et al. FAIR: Quality-aware federated learning with precise user incentive and model aggregation[C]. Proceedings of the IEEE INFOCOM 2021-IEEE Conference on Computer Communications, Vancouver, Canada, 2021: 1–10. doi: 10.1109/INFOCOM42981.2021.9488743. [30] LITJENS G, TOTH R, VAN DE VEN W, et al. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge[J]. Medical Image Analysis, 2014, 18(2): 359–373. doi: 10.1016/j.media.2013.12.002. [31] LEMAÎTRE G, MARTÍ R, FREIXENET J, et al. Computer-aided detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: A review[J]. Computers in Biology and Medicine, 2015, 60: 8–31. doi: 10.1016/j.compbiomed.2015.02.009. [32] BLOCH N, MADABHUSHI A, HUISMAN H, et al. NCI-ISBI 2013 challenge: Automated segmentation of prostate structures. The cancer imaging archive[EB/OL]. http://doi.org/10.7937/K9/TCIA.2015.zF0vlOPv, 2015. [33] MCMAHAN B, MOORE E, RAMAGE D, et al. Communication-efficient learning of deep networks from decentralized data[C]. Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, USA, 2017: 1273–1282. [34] LIANG Xiaoxiao, LIN Yiqun, FU Huazhu, et al. RSCFed: Random sampling consensus federated semi-supervised learning[C]. Proceedings of 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 10144–10153. doi: 10.1109/CVPR52688.2022.00991. [35] MA Yuxi, WANG Jiacheng, YANG Jing, et al. Model-heterogeneous semi-supervised federated learning for medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2024, 43(5): 1804–1815. doi: 10.1109/TMI.2023.3348982. (查阅网上资料,本条文献与第7条文献重复,请确认). [36] XIAO Yunpeng, ZHANG Qunqing, TANG Fei, et al. Cycle-fed: A double-confidence unlabeled data augmentation method based on semi-supervised federated learning[J]. IEEE Transactions on Mobile Computing, 2024, 23(12): 11014–11028. doi: 10.1109/TMC.2024.3388731. [37] XI Yuan, LI Qiong, and MAO Haokun. Federated semi-supervised learning via globally guided pseudo-labeling: A robust approach for label-scarce scenarios[J]. Expert Systems with Applications, 2025, 294: 128667. doi: 10.1016/j.eswa.2025.128667. -





 下载:
下载:
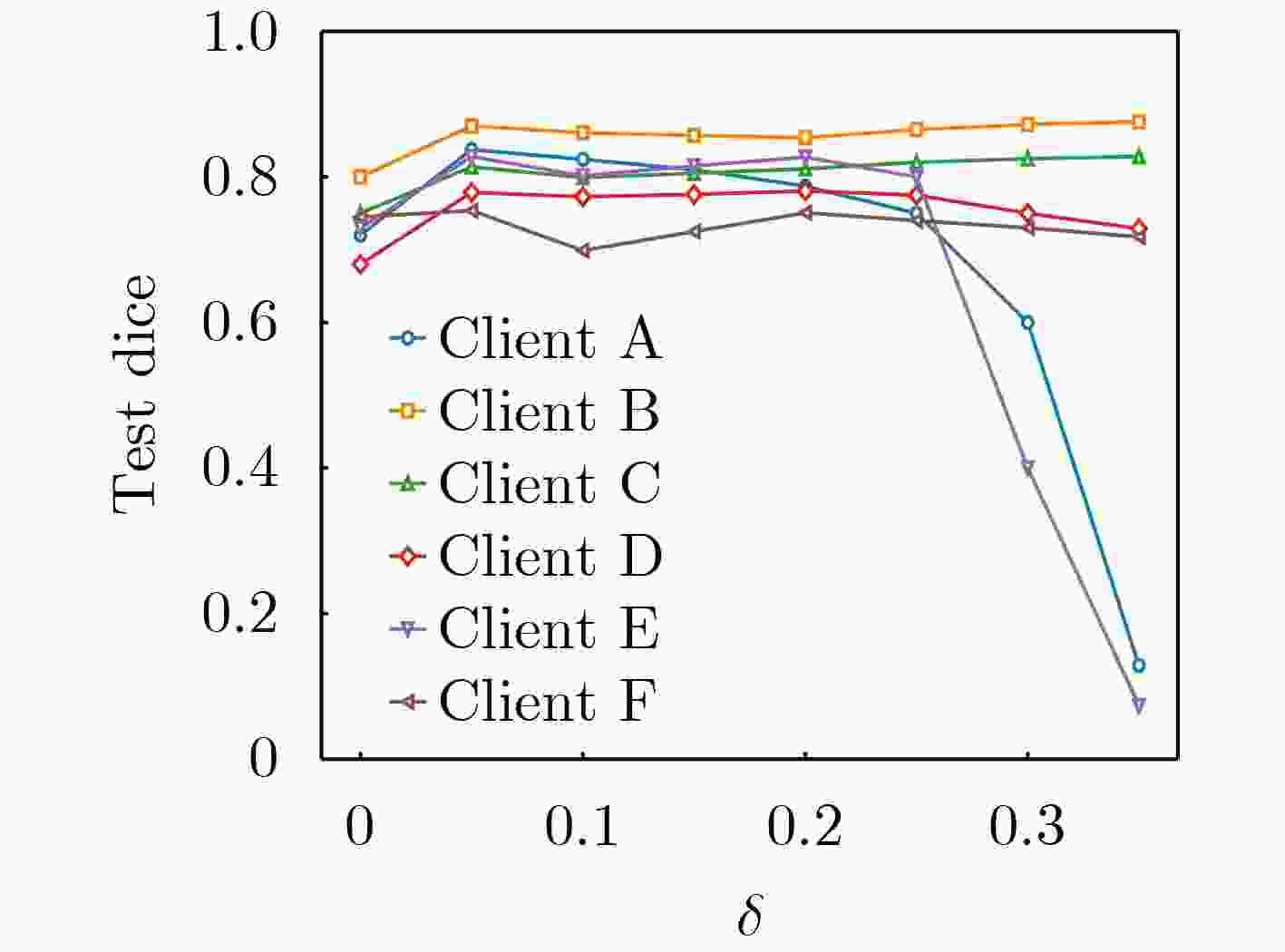
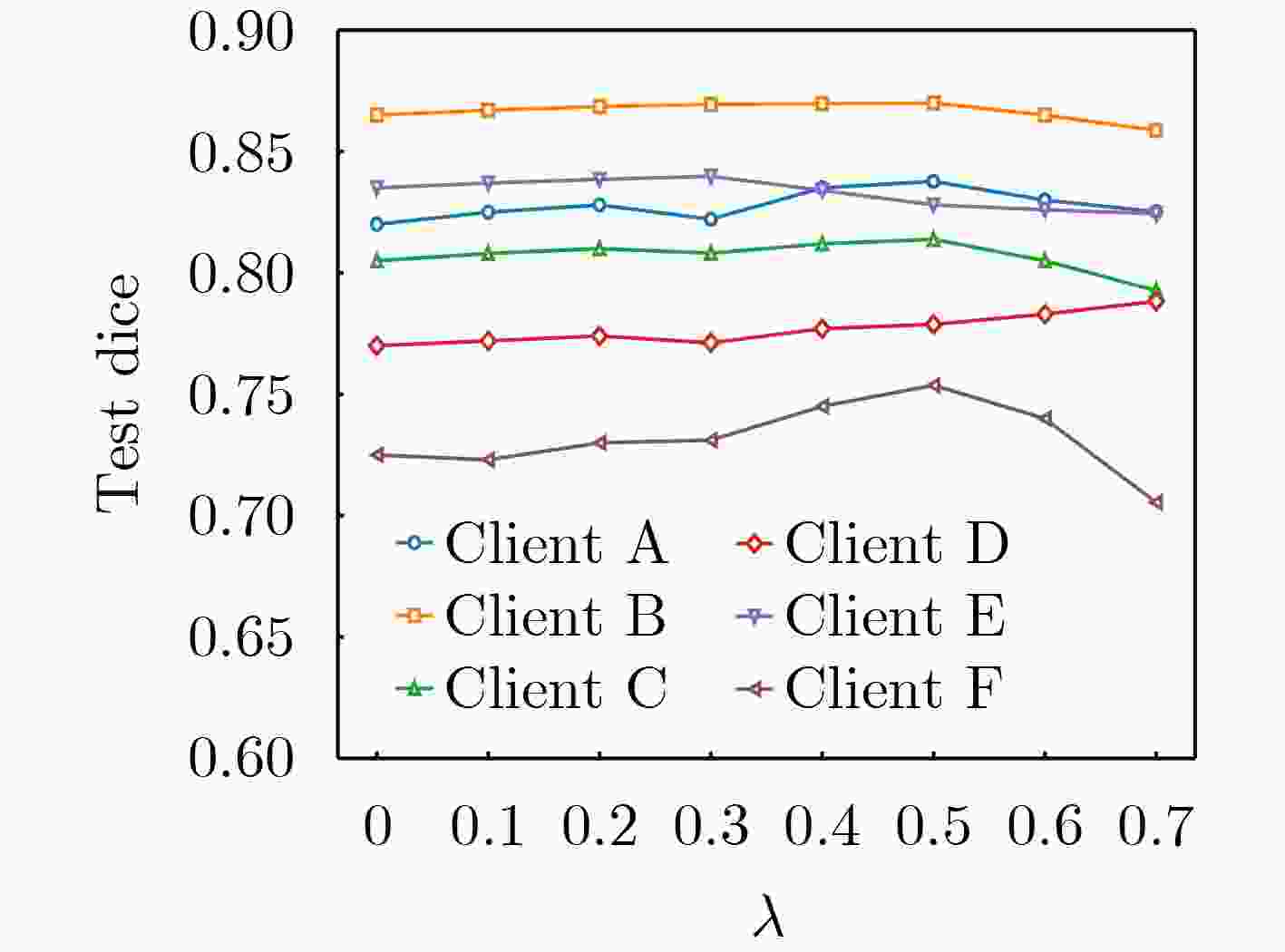


 下载:
下载:
