Component Placement Algorithm Considering Reagent Type Differences in Cell Reuse for FPVA Biochips
-
摘要: 作为新型流式微流控生物芯片,完全可编程阀门阵列(FPVA)生物芯片拥有出色的灵活性和可编程性,能够满足多种复杂的实验需求。而作为架构综合的重要阶段,FPVA组件布局会影响包括生物测定完成时间、流体运输路径总长度和交叉污染程度在内的多个性能指标。此外,单元复用是FPVA芯片灵活性和可编程性的重要表现。然而,现有的FPVA组件布局相关研究工作均没有考虑单元复用过程中试剂种类的差异对这些重要指标的影响。为此,该文聚焦于FPVA组件布局过程中单元复用的试剂种类差异问题,提出了基于深度强化学习的考虑单元复用中试剂种类差异的FPVA组件布局算法。首先,通过考虑试剂之间的差异和组件重叠对交叉污染的影响,设计了单元复用复杂度和含有相同试剂组件平均距离两个指标,分别用于量化组件布局方案中各个单元的单元复用复杂程度和含有相同试剂组件的聚集程度。其次,引入了包括组件布局区域限制约束和并发组件不重叠约束在内的约束条件,这些约束确保组件布局方案的合法性。最后,通过设计奖励函数最小化单元复用复杂度和减小含有相同种类试剂的组件间距离,从而达成最小化最终组件布局方案的交叉污染程度、流体运输路径总长度和生物测定完成时间的目标。通过仿真实验,对所提出的组件布局算法进行了评估。实验结果表明,所提算法能够得到高质量的考虑单元复用中试剂种类差异的FPVA组件布局方案,验证了所提算法的有效性。Abstract:
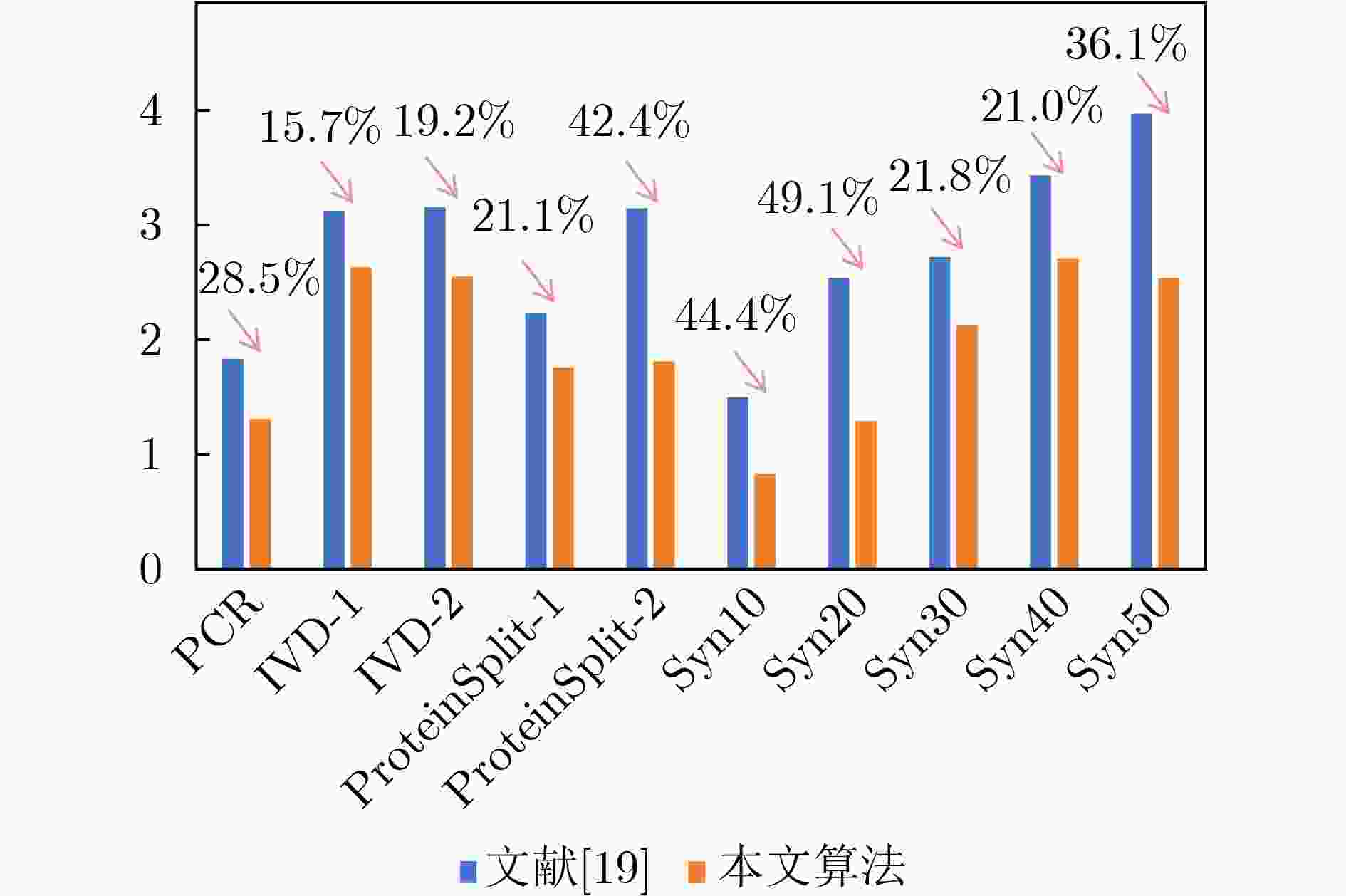
Objective Fully Programmable Valve Array (FPVA) biochips, as a novel type of flow-based microfluidic biochips, offer exceptional flexibility and programmability, enabling them to meet diverse and complex experimental requirements. Among the various stages of FPVA architectural synthesis, component placement design plays a critical role, as it directly impacts several key performance metrics, including the completion time of the bioassay, the total length of the fluid transport path, and the degree of cross-contamination. As an important manifestation of FPVA's flexibility and programmability, cell reuse requires special attention during the component placement process. However, existing FPVA component placement research has largely overlooked the impact of reagent type differences in cell reuse on these performance indicators. Methods This paper presents a component placement algorithm for FPVA biochip that takes into account the differences in reagent types during cell reuse. First, by considering the influence of reagent-type differences and component overlap on cross-contamination, this proposed algorithm introduces a cell reuse complexity metric to quantify the complexity of cell reuse in component placement solutions. Second, the proposed algorithm incorporates constraints, including component placement area restrictions and non-overlapping constraints for concurrent components, to ensure the validity of component placement solutions. Finally, the proposed algorithm optimizes the reward function to minimize cell reuse complexity and reduce the distance between components using the same type of reagent. The proposed algorithm aims to minimize cross-contamination, the total length of the fluid transport path, and the completion time of the bioassay in the final component placement. Results and Discussions The proposed algorithm conducts simulation experiments on benchmark FPVA instances with varying chip sizes and functional requirements, comparing the proposed algorithm with existing related methods. Results show that the proposed algorithm achieves an average reduction of 34.2% in cell reuse complexity, 2.8% in the completion time of the bioassay, and 9.2% in the total length of the fluid transport path ( Table 2 ), while also reducing the reagent-aware distance metric by an average of 29.9% (Fig. 6 ). The decision trajectories of the learning agent demonstrate clear spatial regularity, highlighting the model's awareness of global placement structures.Conclusions This paper is the first to investigate the component placement problem of FPVA biochips considering reagent type differences in cell reuse. The main contributions and findings are as follows: (1) A cell reuse complexity metric is designed to measure the degree of cell reuse in component placement. (2) The FPVA component placement task is effectively modeled as a Markov decision process, enabling the use of double deep Q-networks to learn efficient and safe component placement policies. (3) Compared with existing work, the proposed model significantly improves the biochemical assay performance and reliability of FPVA biochips. -
Key words:
- Microfluidic Biochip /
- FPVA /
- Cell Reuse /
- Reagent Type /
- Component Placement
-
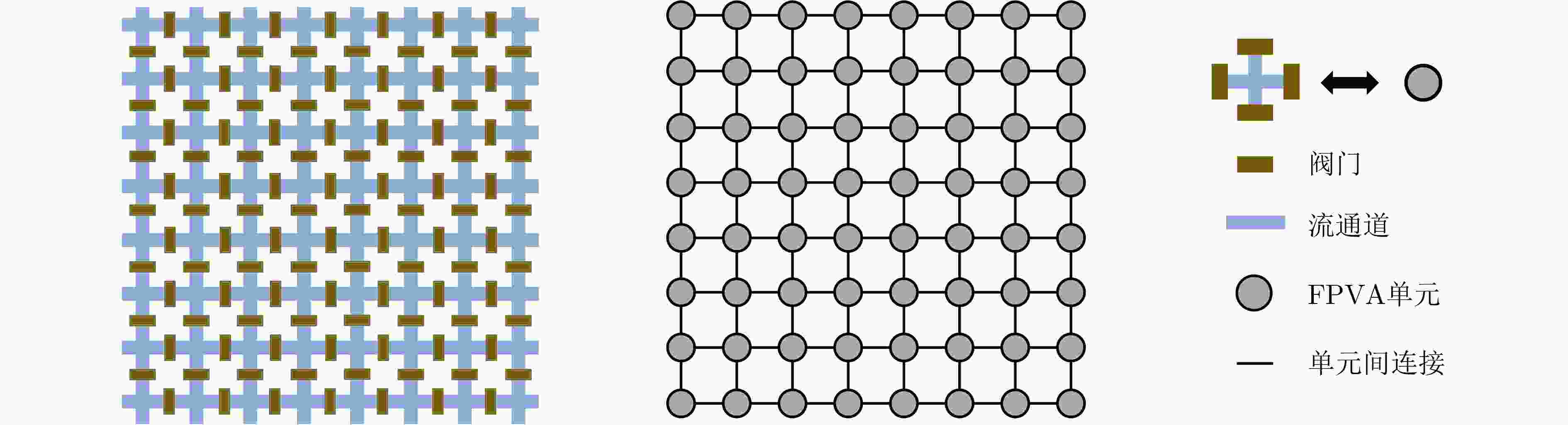
图 2 FPVA生物芯片建模[19]
图 6 本文算法与文献[19]的RAD对比图
表 1 测试用例具体信息
测试用例 试剂种类数 输入流体数 输入通道分配 输出流体数 混合操作数 FPVA尺寸 PCR 3 8 2-2-4 1 7 8×8 IVD1 4 12 3×4 6 6 8×8 IVD2 6 18 2-2-3×3-5 9 9 10×10 ProteinSplit1 5 14 1-2-3-4-4 2 12 10×10 ProteinSplit2 11 32 1-2×4-3×3-4-5-5 4 23 13×13 Synthetic1 2 5 2-3 1 4 8×8 Synthetic2 5 13 1-1-3-3-5 1 12 10×10 Synthetic3 6 18 1-2-3-3-4-5 1 17 12×12 Synthetic4 8 22 1-2-3×5-4 1 21 13×13 Synthetic5 9 27 1-1-2-2-3-3-4-5-6 1 26 13×13 表 2 本文算法与文献[19]中算法的对比结果
测试用例 总单元复用复杂度 生物测定完成时间(s) 流体运输路径总长度(mm) 文献[19] 本文算法 优化/% 文献[19] 本文算法 优化/% 文献[19] 本文算法 优化/% PCR 8.5 3.7 56.5 17.0 16.3 4.1 30 28 6.7 IVD1 15.0 9.0 40.0 4.5 4.3 4.4 46 43 6.5 IVD2 46.0 32.0 30.4 5.4 4.9 9.3 82 71 13.4 ProteinSplit1 108.4 104.0 4.1 30.2 29.9 1.0 83 80 3.6 ProteinSplit2 2403.9 1317.7 45.2 38.4 37.6 2.1 226 198 12.4 Synthetic1 4.0 1.0 75.0 12.8 12.7 0.8 28 25 10.7 Synthetic2 73.2 65.2 10.9 21.2 20.9 1.4 78 72 7.7 Synthetic3 187.0 135.6 27.5 25.7 25.2 1.9 127 112 11.8 Synthetic4 287.9 179.3 37.7 30.4 29.6 2.6 199 195 2.0 Synthetic5 502.9 429.5 14.6 30.3 30.1 0.7 245 202 17.6 平均 34.2 2.8 9.2 表 3 本文算法与文献[24]中算法的对比结果
测试用例 总单元复用复杂度 生物测定完成时间(s) 流体运输路径总长度(mm) 文献[24] 本文算法 优化/% 文献[24] 本文算法 优化/% 文献[24] 本文算法 优化/% PCR 8.2 3.7 54.9 16.8 16.3 3.0 29 28 3.4 IVD1 14.5 9.0 37.9 4.4 4.3 2.3 45 43 4.4 IVD2 44.5 32.0 28.1 5.3 4.9 7.5 80 71 11.3 ProteinSplit1 107.0 104.0 2.8 30.0 29.9 0.3 82 80 2.4 ProteinSplit2 2380.0 1317.7 44.6 38.2 37.6 1.6 224 198 11.6 Synthetic1 3.9 1.0 74.4 12.7 12.7 0.0 27 25 7.4 Synthetic2 72.0 65.2 9.4 21.0 20.9 0.5 77 72 6.5 Synthetic3 184.0 135.6 26.3 25.5 25.2 1.2 125 112 10.4 Synthetic4 284.0 179.3 36.9 30.2 29.6 2.0 197 195 1.0 Synthetic5 498.0 429.5 13.8 30.2 30.1 0.3 242 202 16.5 平均 32.9 1.9 7.5 -
[1] MINHASS W H, POP P, MADSEN J, et al. Architectural synthesis of flow-based microfluidic large-scale integration biochips[C]. Proceedings of the International Conference on Compilers, Architectures and Synthesis for Embedded Systems, Tampere, Finland, 2012: 181–190. doi: 10.1145/2380403.2380437. [2] MINHASS W H, POP P, and MADSEN J. Synthesis of biochemical applications on flow-based microfluidic biochips using constraint programming[C]. Proceedings of the Symposium on Design, Test, Integration and Packaging of MEMS/MOEMS, Cannes, France, 2012: 37–41. (查阅网上资料, 未找到doi信息, 请确认). [3] 朱予涵, 刘博文, 黄兴, 等. 完全可编程阀门阵列生物芯片下容错导向的高阶综合算法[J]. 电子与信息学报, 2024, 46(11): 4141–4150. doi: 10.11999/JEIT240049.ZHU Yuhan, LIU Bowen, HUANG Xing, et al. Fault-tolerance-oriented high-level synthesis algorithm for fully programmable valve array biochips[J]. Journal of Electronics & Information Technology, 2024, 46(11): 4141–4150. doi: 10.11999/JEIT240049. [4] MCDANIEL J, PARKER B, and BRISK P. Simulated annealing-based placement for microfluidic large scale integration (mLSI) chips[C]. Proceedings of the 22nd International Conference on Very Large Scale Integration, Playa del Carmen, Mexico, 2014: 1–6. (查阅网上资料, 未找到doi信息, 请确认). [5] HU Kai, HO T Y, and CHAKRABARTY K. Wash optimization and analysis for cross-contamination removal under physical constraints in flow-based microfluidic biochips[J]. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 2016, 35(4): 559–572. doi: 10.1109/TCAD.2015.2488485. [6] GRIMMER A, WANG Qin, YAO Hailong, et al. Close-to-optimal placement and routing for continuous-flow microfluidic biochips[C]. Proceedings of the 22nd Asia and South Pacific Design Automation Conference, Chiba, Japan, 2017: 530–535. doi: 10.1109/ASPDAC.2017.7858377. [7] CRITES B, KONG K, and BRISK P. Diagonal component expansion for flow-layer placement of flow-based microfluidic biochips[J]. ACM Transactions on Embedded Computing Systems, 2017, 16(5s): 126. doi: 10.1145/3126529. [8] CHEN Zhisheng, HUANG Xing, GUO Wenzhong, et al. Physical synthesis of flow-based microfluidic biochips considering distributed channel storage[C]. Proceedings of the Design, Automation & Test in Europe Conference & Exhibition, Florence, Italy, 2019: 1525–1530. doi: 10.23919/DATE.2019.8715269. [9] 王钦. 微流控生物芯片流体与控制协同物理设计算法研究[D]. [博士论文], 清华大学, 2018. doi: 10.27266/d.cnki.gqhau.2018.000723.WANG Qin. Research on control-fluidic physical CoDesign algorithm for microfluidic biochips[D]. [Ph. D. dissertation], Tsinghua University, 2018. doi: 10.27266/d.cnki.gqhau.2018.000723. [10] 朱予涵, 黄鸿斌, 林泓星, 等. 连续微流控生物芯片下基于序列对的流层物理设计算法[J]. 计算机辅助设计与图形学学报, 2022, 34(4): 535–544. doi: 10.3724/SP.J.1089.2022.19445.ZHU Yuhan, HUANG Hongbin, LIN Hongxing, et al. Sequence-pair-based flow-layer physical design algorithm for continuous-flow microfluidic biochips[J]. Journal of Computer-Aided Design & Computer Graphics, 2022, 34(4): 535–544. doi: 10.3724/SP.J.1089.2022.19445. [11] 刘耿耿, 叶正阳, 朱予涵, 等. 连续微流控生物芯片下一种多阶段启发式的流层物理协同设计算法[J]. 电子与信息学报, 2023, 45(9): 3401–3409. doi: 10.11999/JEIT221155.LIU Genggeng, YE Zhengyang, ZHU Yuhan, et al. A multi-stage heuristic flow-layer physical codesign algorithm for continuous-flow microfluidic biochips[J]. Journal of Electronics & Information Technology, 2023, 45(9): 3401–3409. doi: 10.11999/JEIT221155. [12] FIDALGO L M and MAERKL S J. A software-programmable microfluidic device for automated biology[J]. Lab on a Chip, 2011, 11(9): 1612–1619. doi: 10.1039/C0LC00537A. [13] TSENG T M, LI Bing, HO T Y, et al. Reliability-aware synthesis for flow-based microfluidic biochips by dynamic-device mapping[C]. Proceedings of the 52nd ACM/EDAC/IEEE Design Automation Conference, San Francisco, USA, 2015: 1–6. doi: 10.1145/2744769.2744899. [14] TSENG T M, LI Bing, LI Mengchu, et al. Reliability-aware synthesis with dynamic device mapping and fluid routing for flow-based microfluidic biochips[J]. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 2016, 35(12): 1981–1994. doi: 10.1109/TCAD.2016.2547902. [15] CHOUDHARY G, PAL S, KUNDU D, et al. Transport-free module binding for sample preparation using microfluidic fully programmable valve arrays[C]. Proceedings of the Design, Automation & Test in Europe Conference & Exhibition, Grenoble, France, 2020: 1335–1338. doi: 10.23919/DATE48585.2020.9116370. [16] DATTA P, CHAKRABORTY A, and PAL R K. Design optimization for programmable microfluidic devices integrating contamination removal and capacity-wastage-aware washing[J]. IETE Journal of Research, 2020, 66(6): 781–796. doi: 10.1080/03772063.2020.1811784. [17] KUNDU D, GIRI J, MARUYAMA S, et al. Fluid-to-cell assignment and fluid loading on programmable microfluidic devices for bioprotocol execution[J]. Integration, 2021, 78: 95–109. doi: 10.1016/j.vlsi.2020.12.004. [18] YU H C, LIN Y H, CHEN Zhiyang, et al. Contamination-aware synthesis for programmable microfluidic devices[J]. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 2022, 41(11): 5016–5029. doi: 10.1109/TCAD.2021.3134892. [19] LIU Genggeng, ZHU Yuhan, GUO Wenzhong, et al. Fault-tolerance-oriented physical design for fully programmable valve array biochips[C]. Proceedings of the 60th ACM/IEEE Design Automation Conference, San Francisco, USA, 2023: 1–6. doi: 10.1109/DAC56929.2023.10247720. [20] KUMAR M, KHAN A K, ROY S, et al. Accelerating fluid loading in sample preparation with fully programmable valve arrays[C]. Proceedings of the 37th International Conference on VLSI Design and 23rd International Conference on Embedded Systems (VLSID), Kolkata, India, 2024: 402–407. doi: 10.1109/VLSID60093.2024.00073. [21] ZHU Yuhan, LIU Genggeng, GUO Wenzhong, et al. FTCD: Fault-tolerant co-design of flow and control layers for fully programmable valve array biochips[J]. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 2025, 44(7): 2669–2682. doi: 10.1109/TCAD.2025.3525615. [22] CAI Huayang, LIU Genggeng, GUO Wenzhong, et al. Adaptive control-logic routing for fully programmable valve array biochips using deep reinforcement learning[C]. Proceedings of the 29th Asia and South Pacific Design Automation Conference, Incheon, Korea, 2024: 564–569. doi: 10.1109/ASP-DAC58780.2024.10473962. [23] HUANG Xing, CAI Huayang, GUO Wenzhong, et al. Control-logic synthesis of fully programmable valve array using reinforcement learning[J]. IEEE Transactions on Computer-Aided Design of Integrated Circuits and Systems, 2024, 43(1): 277–290. doi: 10.1109/TCAD.2023.3309740. [24] MNIH V, KAVUKCUOGLU K, SILVER D, et al. Playing Atari with deep reinforcement learning[J]. arXiv preprint arXiv: 1312.5602, 2013. doi: 10.48550/arXiv.1312.5602. (查阅网上资料,不确定本条文献的格式和类型,请确认). -





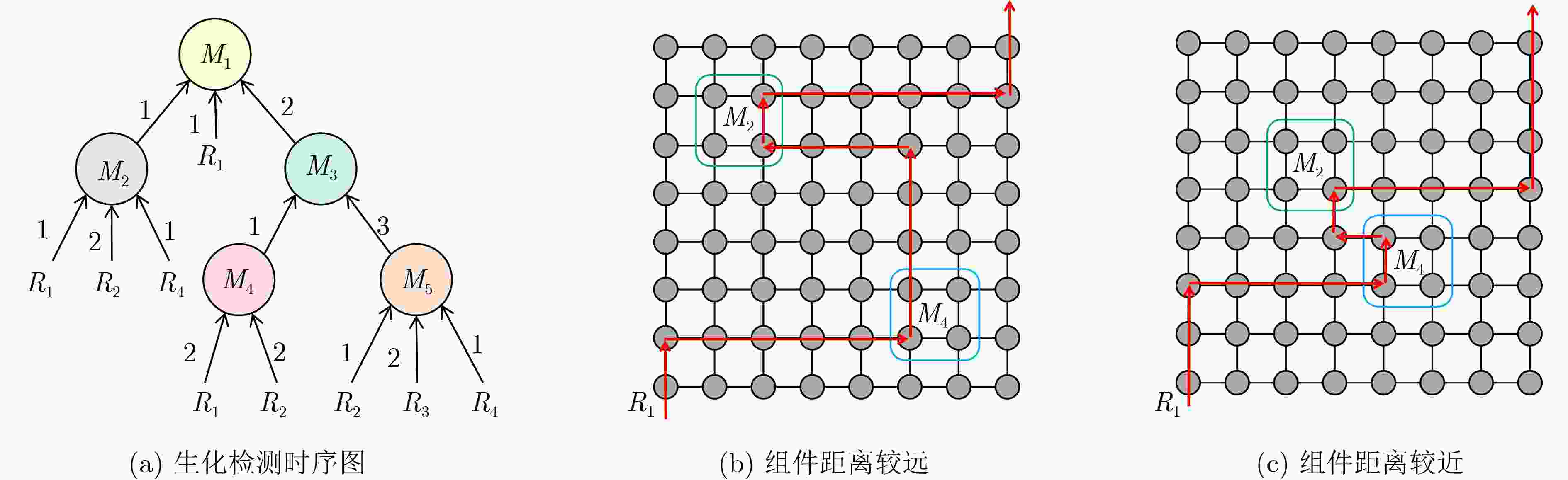
 下载:
下载:
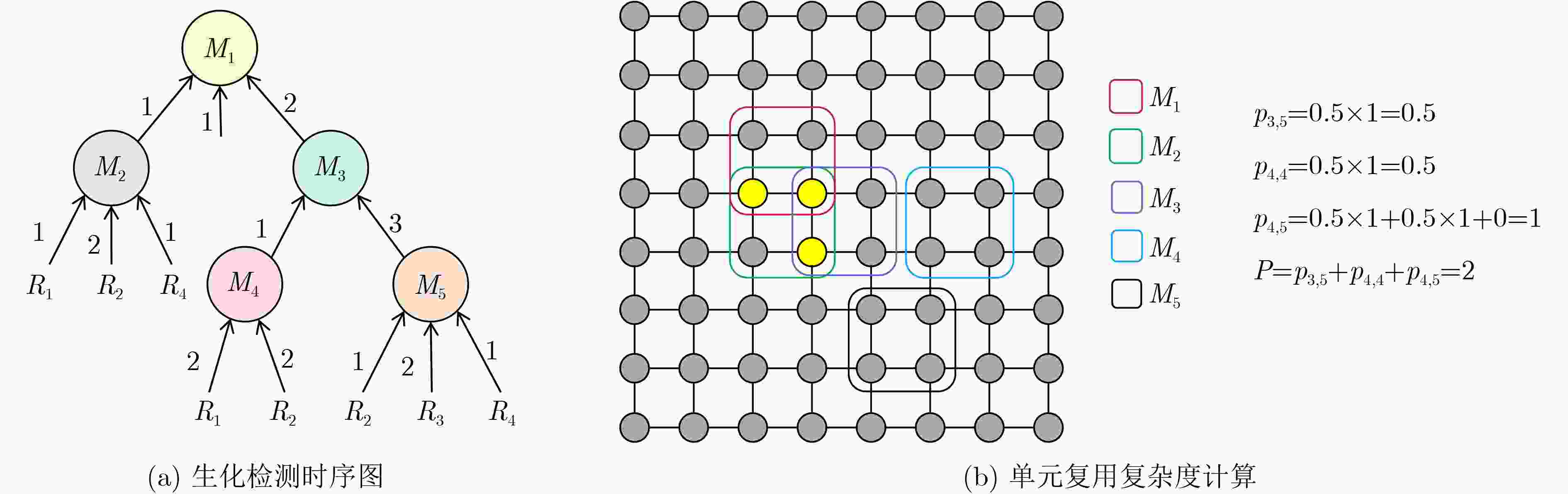



 下载:
下载:
