Hierarchical Fusion Multi-Instance Learning for Weakly Supervised Pathological Image Classification
-
摘要: 病理图像分类对于癌症诊断至关重要,但现有方法存在依赖随机采样、多层级信息利用不足等问题。为此,该文提出一种层级融合多实例学习方法。首先,对病理图像的不同层级进行切分并用ResNet-50提取特征;然后,针对伪包标签不准确及背景噪声问题,提出基于注意力评价函数的伪包划分方法,利用门控注意力对低分辨率特征进行重要性评估,依据得分将特征划分为低分辨率伪包,并通过索引映射得到对应高分辨率伪包;最后,针对多层级信息利用不足的问题设计两阶段分类模型,第一阶段对低分辨率伪包进行初步分类,并依据预测置信度筛选出高判别性的关键区域及其对应的高分辨率特征;第二阶段通过交叉注意力机制,将筛选出的低分辨率特征与对应的高分辨率特征进行深度融合,随后将其与经过门控注意力聚合的高分辨率伪包特征进行拼接,以利用局部细节结合全局信息进行分析。在训练过程中,采用双分支交叉熵损失函数,联合优化低分辨率初步分类任务与高分辨率最终分类任务。实验使用了两个公开数据集Camelyon16、TCGA-LUNG及一个私有皮肤癌数据集NBU-Skin对模型进行测试,结果表明,该方法在多中心公开数据集和私有数据集上性能均优于CLAM、TransMIL等算法,其中在NBU-Skin数据集上五折交叉验证的平均准确率达到90.5%,平均AUC达到0.976。此外,该方法在跨病种、跨中心数据中表现稳定,为癌症病理的人工智能诊断提供了新的思路。Abstract:
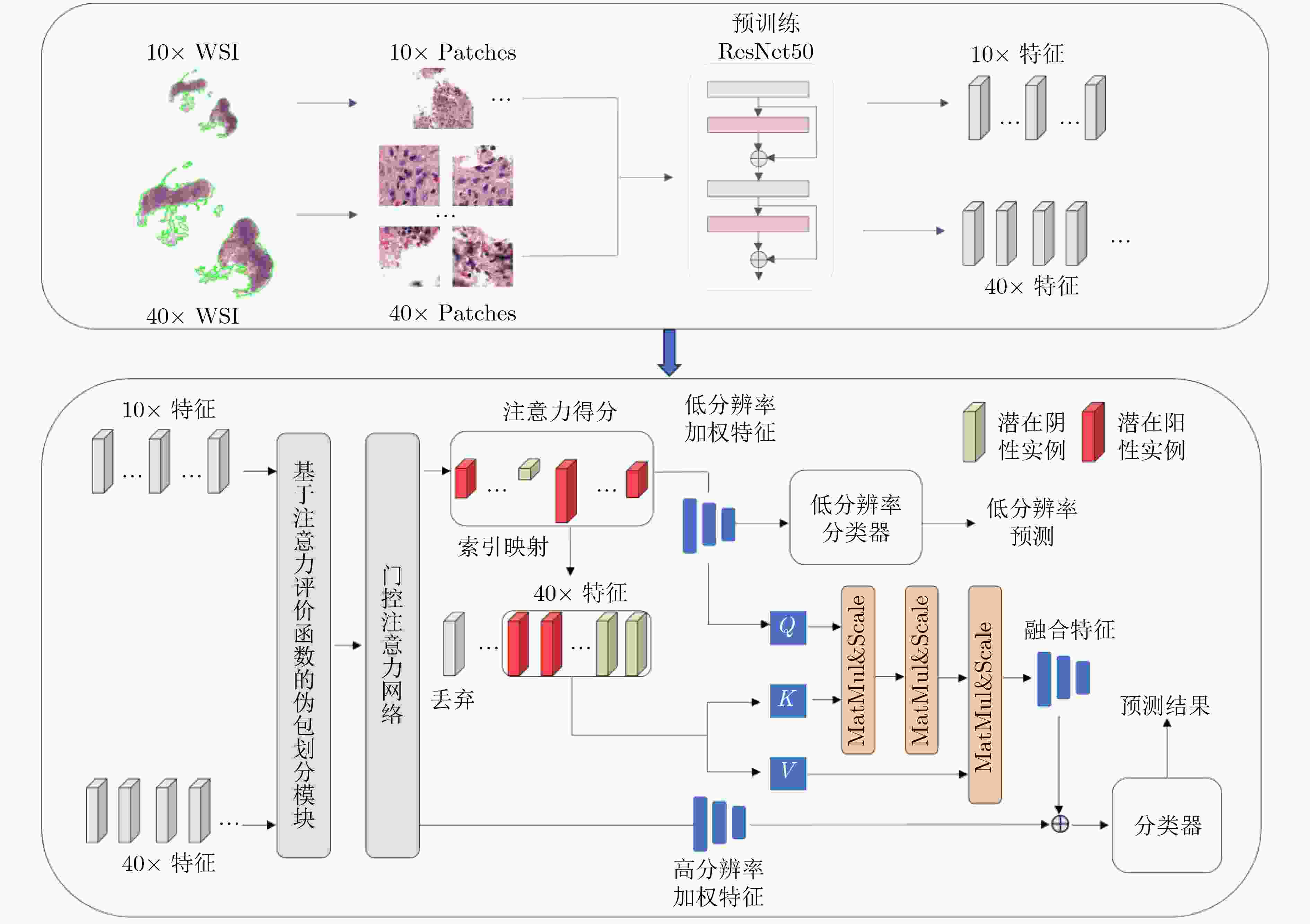
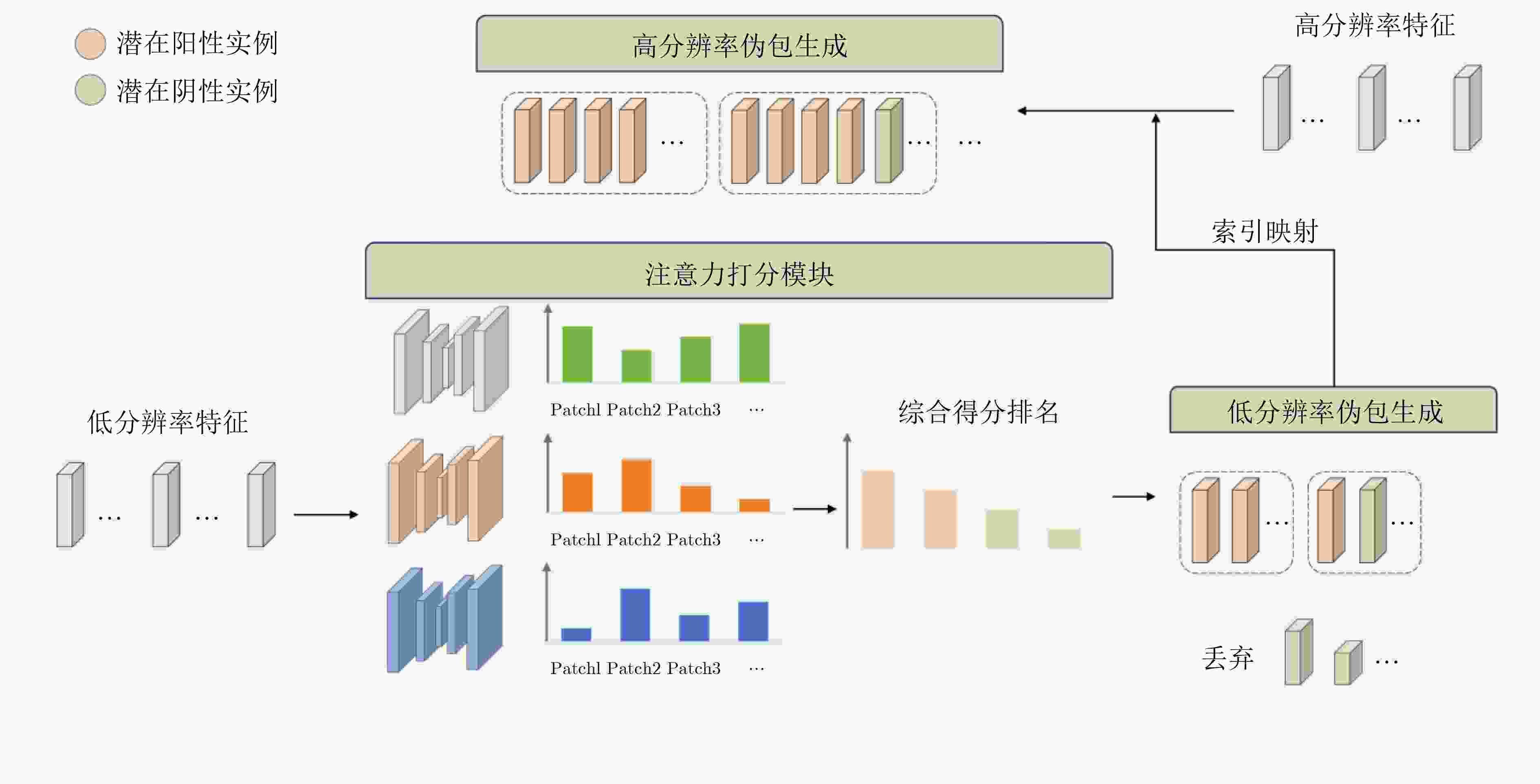
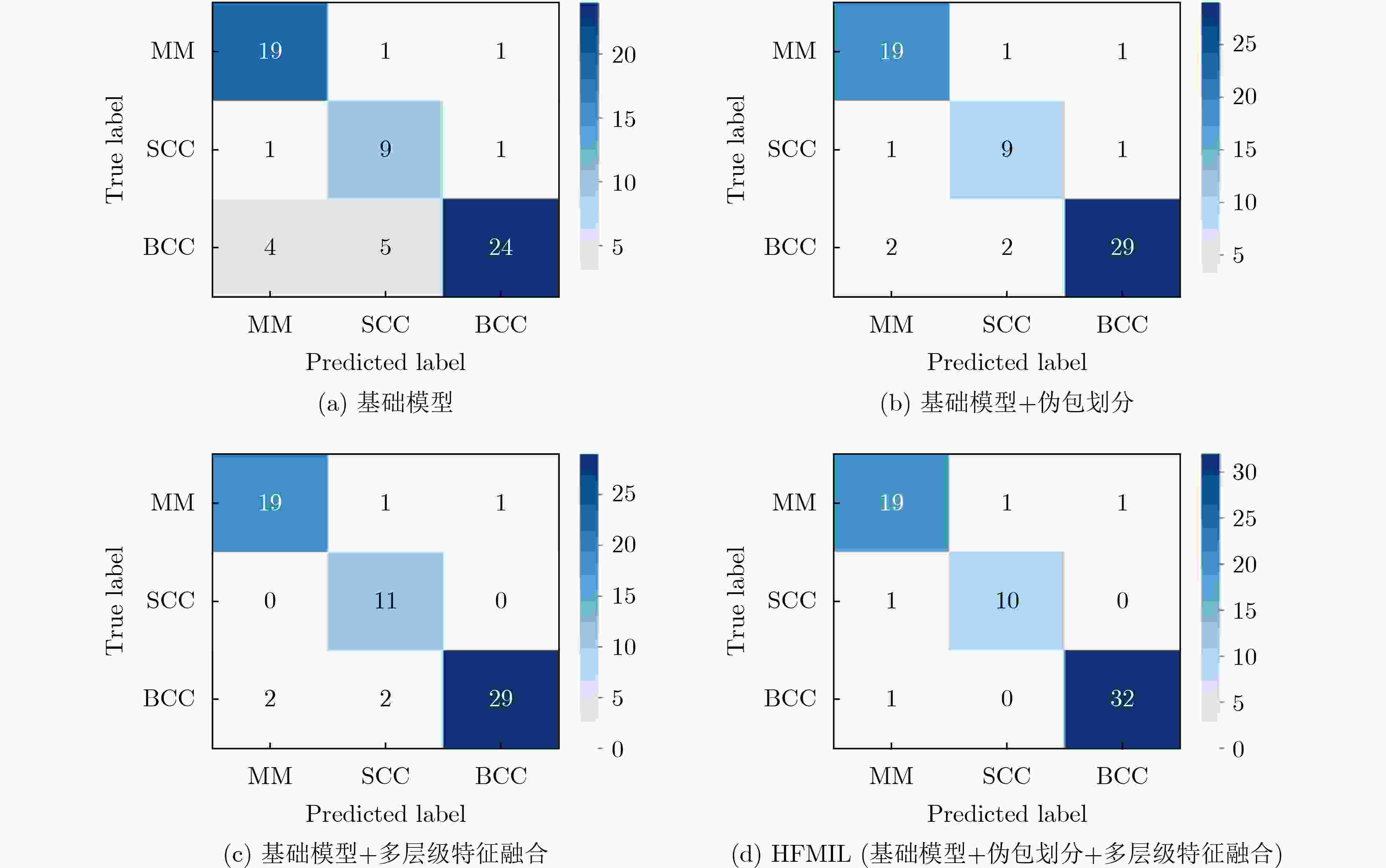
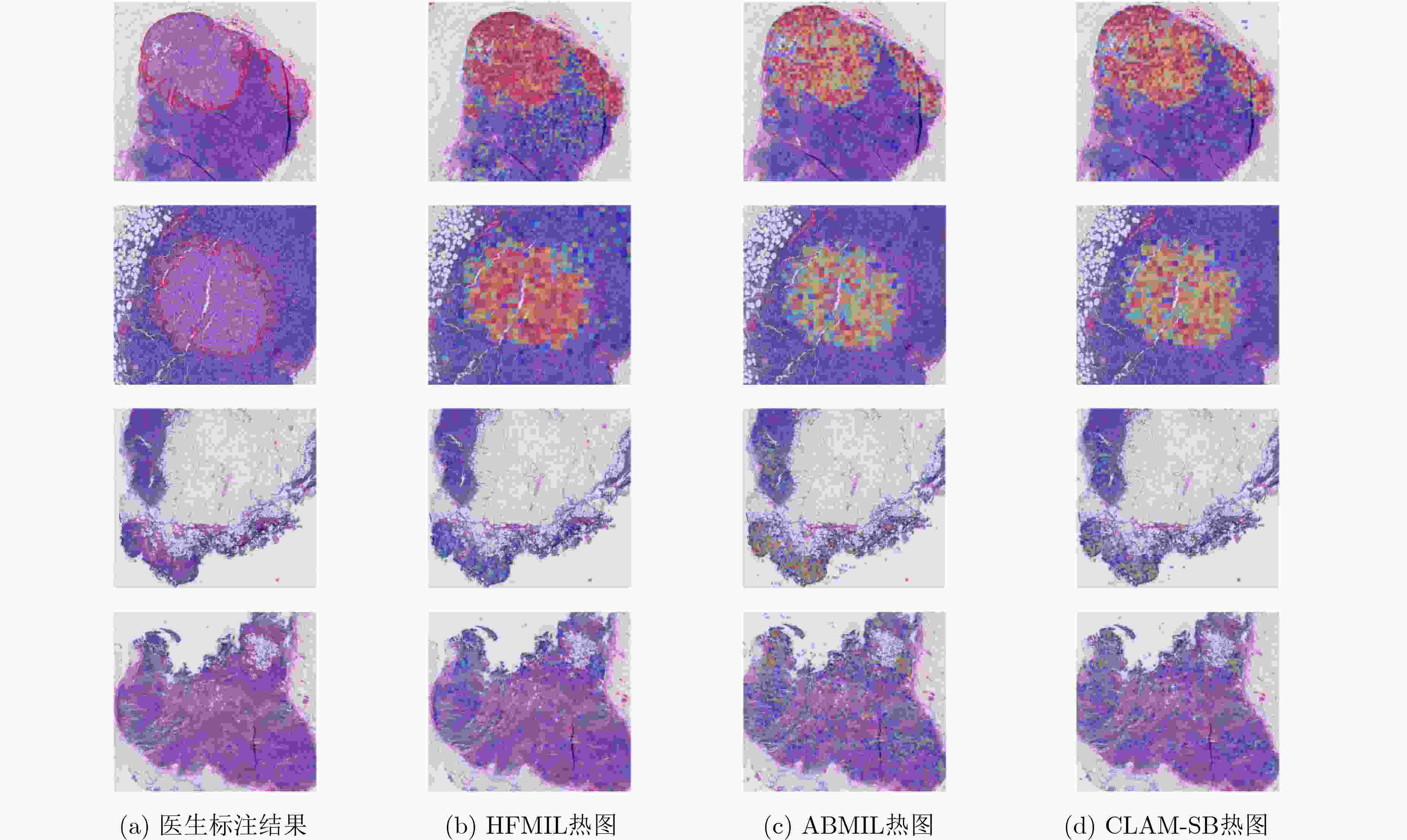
Objective As the mortality rates of cancer in China continue to rise, the significance of pathological image classification in cancer diagnosis is increasingly recognized. Pathological images are characterized by a multi-level structure. However, most existing methods primarily focus on the highest resolution of pathological images or employ simple feature concatenation strategies to fuse multi-scale information, failing to effectively utilize the multi-level information inherent in these images. Furthermore, existing methods typically employ random pseudo-bag division strategies to address the challenge of high-resolution pathological images. However, due to the sparsity of cancerous regions in positive slides, such random sampling often results in incorrect pseudo-labels and low signal-to-noise ratios, thereby posing additional challenges to classification accuracy. To address these issues, this study proposes a Hierarchical Fusion Multi-Instance Learning (HFMIL) method, integrating multi-level feature fusion with a pseudo-bag division strategy based on an attention evaluation function. It is designed to enhance the accuracy and interpretability of pathological image classification, thereby providing a more effective tool for clinical diagnosis. Methods A weakly supervised classification method based on a multi-level model is proposed in this study to leverage the multi-level characteristics of pathological images, thereby enhancing the performance of cancer pathological image classification. The proposed method is composed of three core steps. Initially, multi-level feature extraction is performed. Blank areas are removed from pathological images, and low-resolution images are segmented into image patches and then mapped to their corresponding high-resolution patches through index mapping. Semantic features are extracted to capture multi-level information, including low-resolution tissue structures and high-resolution cellular details. Subsequently, a pseudo-bag division method based on an attention evaluation function is employed. Classification scores for each image patch are computed through class activation mapping (CAM) to assess the importance of low-resolution features. Patches are ranked by scores, and potential positive features are selected to form pseudo-bags, while low-scoring features are discarded to ensure that pseudo-bags contain information relevant to pseudo-labels. Corresponding high-resolution pseudo-bags are then generated via feature index mapping, effectively addressing the issues of incorrect pseudo-labels and low signal-to-noise ratios. Finally, a two-stage classification model is developed. In the first stage, low-resolution pseudo-bags are aggregated using a gated attention mechanism for preliminary classification. In the second stage, a cross-attention mechanism is employed to fuse the most contributory low-resolution features with their corresponding high-resolution counterparts. The fused features are then concatenated with the aggregated high-resolution pseudo-bags to form a comprehensive image-level feature representation, which is input into a classifier for final prediction. Model training is conducted using a two-stage loss function, combining cross-entropy losses from low-resolution classification and overall classification to ensure effective integration of multi-level information. The method is experimentally validated on three pathological image datasets, demonstrating its effectiveness in weakly supervised pathological image classification tasks. Results and Discussions The proposed method is compared with several state-of-the-art weakly supervised classification methods, including ABMIL, CLAM, TransMIL, and DTFD. Evaluations are conducted on three pathological image datasets: the publicly available Camelyon16 and TCGA-LUNG datasets, and a private skin cancer dataset, NBU-Skin. Experimental results indicate that the proposed method achieves significant performance improvements on the test sets. On the Camelyon16 dataset, a classification accuracy of 88.3% and an AUC value of 0.979 are obtained ( Table 2 ). On the TCGA-LUNG dataset, a classification accuracy of 86.0% and an AUC value of 0.931 are obtained (Table 2 ), surpassing comparative methods. On the NBU-Skin dataset, a classification accuracy of 90.5% and an AUC value of 0.976 are achieved for multi-classification tasks (Table 2 ).To further validate the effectiveness of the proposed approach, ablation studies are conducted to assess the necessity of the multi-level feature fusion and pseudo-bag division modules. The results demonstrate that the combination of these modules enhances classification performance. For instance, on the skin cancer dataset, the removal of the pseudo-bag division module was observed to reduce classification accuracy from 93.8% to 90.7%, and the subsequent removal of the multi-level feature fusion module further reduced it to 80.0% (Table 3 ). These findings collectively confirm the effectiveness of each component in the method.Conclusions A weakly supervised pathological image classification algorithm is proposed in this study, integrating multi-level feature fusion and an attention-based pseudo-bag division method. This approach effectively leverages multi-level information within pathological images and mitigates challenges related to incorrect pseudo-labels and low signal-to-noise ratios. Experimental results demonstrate that the proposed method outperforms existing approaches in terms of classification accuracy and AUC across three pathological image datasets. The primary contributions include: (1) A multi-level feature extraction and fusion strategy. Unlike existing strategies that primarily focus on the highest resolution or employ simple feature concatenation, this method deeply fuses feature information across different levels via a Cross-Attention Mechanism, effectively utilizing multi-scale information. (2) A pseudo-bag division method based on an attention evaluation function. By scoring features to identify potential positive regions and restructuring training samples via pseudo-bag division, this method not only maximizes the correctness of pseudo-labels through a top-k mechanism but also improves the signal-to-noise ratio by discarding low-scoring background noise. (3) Superior performance over all comparative models. The accuracy of weakly supervised pathological image classification is significantly improved, providing new insights for computer-aided cancer diagnosis. The following future research directions are proposed: (1) optimization of cross-level attention mechanisms; (2) extension of the framework to other medical imaging tasks, such as prognosis prediction or lesion segmentation; (3) designing more efficient feature extraction and fusion methods, and exploring their applications in other disease types and tasks, to better meet clinical needs. -
Key words:
- Pathological Image /
- Multiple Instance Learning /
- Deep Learning /
- Artificial Intelligence
-
1 多层级对应特征提取方法
输入:共输入N张病理图像$ {\{W}_{i}\}_{i=1}^{N} $,其中$ {W}_{i} $代表第i张病理图
像,每张病理图像在低分辨率下有m个图像块预训练网络ResNet50 输出:低分辨率特征集合$ \{{\boldsymbol{X}}_{i,\text{low}}\}_{i=1}^{N} $,其中
$ {\boldsymbol{X}}_{i,\text{low}}=\{{x}_{i,1},{x}_{i,2},\cdots ,{x}_{i,m}\} $高分辨率特征集合$ \{{\boldsymbol{X}}_{i,\text{high}}\}_{i=1}^{N} $,其中
$ {\boldsymbol{X}}_{i,\text{high}}=\{{h}_{i,1},{h}_{i,2},\cdots ,{h}_{i,16m}\} $(1) for i←1 to N do (2) 使用最大类间方差法提取组织区域轮廓并去除空白区域 (3) 将$ {W}_{i} $切分为256×256的图像块,得到第k个图像块的低分
辨率坐标 $ \mathrm{low}\_ {\text{coords}}_{k}=({x}_{k},{y}_{k}) $(4) for k←1 to m do (5) 从$ {W}_{i} $中根据坐标$ \mathrm{low}\_ {\text{coords}}_{k}=({x}_{k},{y}_{k}) $提取图像块
$ {P}_{\mathrm{k},\text{low}} $(6) $ {\boldsymbol{f}}_{\text{low}}\leftarrow \mathrm{ResNet}50({P}_{\mathrm{k},\text{low}}) $ (7) $ {\boldsymbol{X}}_{i,\text{low}}.\mathrm{append}({\boldsymbol{f}}_{\text{low}}) $ (8) 通过${\mathrm{high}\_ \text{coords}}_{k}\mathrm{}=\{({x}_{\mathrm{k}}\times 4+i\times 256,{y}_{\mathrm{k}}\times 4+ $
$ j\times 256)\mid i,j\in \{0{,}1,2{,}3\}\} $计算第k个图像块的高分辨
率坐标$ {\mathrm{high}\_ \text{coords}}_{k}\mathrm{}=({x}_{high},{y}_{high}) $(9) foreach $ ({x}_{high},{y}_{high})\in {\mathrm{high}\_ \text{coords}}_{k} $do (10) 从$ {W}_{i} $中根据坐标$ {high\_ coords}_{\mathrm{k}}=({x}_{high},{y}_{high}) $提
取图像块 $ {P}_{\mathrm{k},\text{high}} $(11) $ {\boldsymbol{f}}_{\text{high}}\leftarrow \mathrm{ResNet}50({P}_{\mathrm{k},\text{high}}) $ (12) $ {\boldsymbol{X}}_{i,\text{high}}.\mathrm{append}({\boldsymbol{f}}_{\text{high}}) $ (13) end foreach (14) end for (15) end for 表 1 数据分布情况
数据集 总数 类别分布 数量 训练集 验证集 测试集 NBU-Skin 326 BCC 166 228 33 65 MM 106 SCC 54 TCGA-LUNG 1053 LUSC 541 738 105 210 LUAD 512 Camelyon16 399 NORMAL 239 243 27 129 TUMOR 160 表 2 模型结果对比
模型 模型推理时间 显存消耗 Camelyon16 NBU-Skin TCGA-LUNG 准确率 AUC 准确率 AUC 准确率 AUC Mean-Pooling[23] 0.229 ms 2148 MB 0.675 0.761 0.755±0.050 0.863±0.114 0.813±0.094 0.881±0.099 Max-Pooling[23] 0.232 ms 2148 MB 0.587 0.599 0.797±0.018 0.881±0.044 0.801±0.031 0.860±0.034 ABMIL[13] 0.468 ms 2276 MB 0.862 0.876 0.838±0.063 0.927±0.089 0.844±0.023 0.919±0.026 Dsmil[17] 0.864 ms 2284 MB 0.836 0.862 0.727±0.027 0.809±0.107 0.783±0.041 0.856±0.040 Dsmil + APBD 0.913 ms 2463 MB 0.850 0.919 0.847±0.039 0.925±0.044 0.844±0.011 0.918±0.016 Dsmil + RankMix[18] 1.176 ms 2949 MB 0.855 0.897 0.823±0.086 0.922±0.042 0.849±0.017 0.917±0.023 Dsmil + ReMix[20] 0.901 ms 3052 MB 0.829 0.905 0.818±0.055 0.908±0.047 0.833±0.037 0.915±0.020 CLAM-SB[14] 0.456 ms 2584 MB 0.806 0.865 0.862±0.070 0.950±0.020 0.834±0.030 0.912±0.034 CLAM-SB + MDDP[24] 0.820 ms 3394 MB 0.872 0.868 0.751±0.083 0.878±0.055 0.841±0.033 0.925±0.042 CLAM-MB[14] 0.841 ms 2584 MB 0.782 0.770 0.865±0.050 0.953±0.032 0.840±0.043 0.912±0.019 TransMIL[15] 6.620 ms 10114 MB 0.858 0.906 0.798±0.100 0.930±0.061 0.819±0.038 0.885±0.030 DTFD(MaxS)[16] 1.833 ms 2140 MB 0.858 0.870 0.859±0.024 0.898±0.012 0.764±0.010 0.837±0.017 DTFD(MaxMinS)[16] 1.911 ms 2336 MB 0.881 0.906 0.792±0.101 0.887±0.074 0.832±0.031 0.907±0.031 DTFD(AFS)[16] 2.052 ms 2130MB 0.881 0.901 0.786±0.101 0.891±0.081 0.849±0.036 0.927±0.019 HFMIL(本文) 2.249 ms 2356 MB 0.883 0.979 0.905±0.030 0.976±0.016 0.860±0.043 0.931±0.026 表 3 消融实验结果
HFMIL消融实验设置 Camelyon16 NBU-Skin TCGA-LUNG 多层级特征融合 伪包划分 准确率 AUC 准确率 AUC 准确率 AUC 0.472 0.420 0.800 0.882 0.785 0.880 √ 0.751 0.668 0.876 0.952 0.823 0.854 √ 0.798 0.782 0.907 0.968 0.880 0.927 √ √ 0.883 0.979 0.938 0.997 0.900 0.966 表 4 top-k参数敏感性实验结果
top-k取值 准确率 AUC 5% 0.907±0.055 0.975±0.013 10% 0.901±0.058 0.971±0.011 15% 0.905±0.030 0.976±0.016 20% 0.900±0.037 0.972±0.015 25% 0.892±0.066 0.966±0.023 -
[1] HAN Bingfeng, ZHENG Rongshou, ZENG Hongmei, et al. Cancer incidence and mortality in China, 2022[J]. Journal of the National Cancer Center, 2024, 4(1): 47–53. doi: 10.1016/j.jncc.2024.01.006. [2] 姜梦琦, 韩昱晨, 傅小龙. 基于人工智能的H-E染色全切片病理学图像分析在肺癌研究中的进展[J]. 中国癌症杂志, 2024, 34(3): 306–315. doi: 10.19401/j.cnki.1007-3639.2024.03.009.JIANG Mengqi, HAN Yuchen, and FU Xiaolong. Research progress on H-E stained whole slide image analysis by artificial intelligence in lung cancer[J]. China Oncology, 2024, 34(3): 306–315. doi: 10.19401/j.cnki.1007-3639.2024.03.009. [3] 王钰萌, 刘振丙, 刘再毅. 隐私保护的联邦弱监督组织病理学亚型分类方法[J/OL]. https://jeit.ac.cn/cn/article/doi/10.11999/JEIT250842, 2025.WANG Yumeng, LIU Zhenbing, and LIU Zaiyi. Privacy-preserving federated weakly-supervised learning for cancer subtyping on histopathology images[J/OL]. https://jeit.ac.cn/cn/article/doi/10.11999/JEIT250842, 2025. [4] 金怀平, 薛飞跃, 李振辉, 等. 基于病理图像集成深度学习的胃癌预后预测方法[J]. 电子与信息学报, 2023, 45(7): 2623–2633. doi: 10.11999/JEIT220655.JIN H P, XUE F Y, LI Z H, et al. Prognostic prediction of gastric cancer based on ensemble deep learning of pathological images[J]. Journal of Electronics & Information Technology, 2023, 45(7): 2623–2633. doi: 10.11999/JEIT220655. [5] FEI Manman, ZHANG Xin, CHEN Dongdong, et al. Whole slide cervical cancer classification via graph attention networks and contrastive learning[J]. Neurocomputing, 2025, 613: 128787. doi: 10.1016/j.neucom.2024.128787. [6] ZHANG Jiawei, SUN Zhanquan, WANG Kang, et al. Prognosis prediction based on liver histopathological image via graph deep learning and transformer[J]. Applied Soft Computing, 2024, 161: 111653. doi: 10.1016/j.asoc.2024.111653. [7] LI Mingze, ZHANG Bingbing, SUN Jian, et al. Weakly supervised breast cancer classification on WSI using transformer and graph attention network[J]. International Journal of Imaging Systems and Technology, 2024, 34(4): e23125. doi: 10.1002/ima.23125. [8] WANG Fuying, XIN Jiayi, ZHAO Weiqin, et al. TAD-graph: Enhancing whole slide image analysis via task-aware subgraph disentanglement[J]. IEEE Transactions on Medical Imaging, 2025, 44(6): 2683–2695. doi: 10.1109/TMI.2025.3545680. [9] WU Kun, JIANG Zhiguo, TANG Kunming, et al. Pan-cancer histopathology WSI pre-training with position-aware masked autoencoder[J]. IEEE Transactions on Medical Imaging, 2025, 44(4): 1610–1623. doi: 10.1109/TMI.2024.3513358. [10] 张印辉, 张金凯, 何自芬, 等. 全局感知与稀疏特征关联图像级弱监督病理图像分割[J]. 电子与信息学报, 2024, 46(9): 3672–3682. doi: 10.11999/JEIT240364.ZHANG Yinhui, ZHANG Jinkai, HE Zifen, et al. Global perception and sparse feature associate image-level weakly supervised pathological image segmentation[J]. Journal of Electronics & Information Technology, 2024, 46(9): 3672–3682. doi: 10.11999/JEIT240364. [11] YAN Rui, LV Zhilong, YANG Zhidong, et al. Sparse and hierarchical transformer for survival analysis on whole slide images[J]. IEEE Journal of Biomedical and Health Informatics, 2024, 28(1): 7–18. doi: 10.1109/JBHI.2023.3307584. [12] MA Yingfan, LUO Xiaoyuan, FU Kexue, et al. Transformer-based video-structure multi-instance learning for whole slide image classification[C]. Proceedings of the 38th AAAI Conference on Artificial Intelligence, Vancouver, Canada, 2024: 14263–14271. doi: 10.1609/aaai.v38i13.29338. [13] ILSE M, TOMCZAK J, and WELLING M. Attention-based deep multiple instance learning[C]. Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 2018: 2127–2136. [14] LU M Y, WILLIAMSON D F K, CHEN T Y, et al. Data-efficient and weakly supervised computational pathology on whole-slide images[J]. Nature Biomedical Engineering, 2021, 5(6): 555–570. doi: 10.1038/s41551-020-00682-w. [15] SHAO Zhuchen, BIAN Hao, CHEN Yang, et al. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification[C]. Proceedings of the 35th International Conference on Neural Information Processing Systems, 2021: 164. (查阅网上资料, 未找到本条文献出版地信息, 请确认). [16] ZHANG Hongrun, MENG Yanda, ZHAO Yitian, et al. DTFD-MIL: Double-tier feature distillation multiple instance learning for histopathology whole slide image classification[C]. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 18780–18790. doi: 10.1109/CVPR52688.2022.01824. [17] LI Bin, LI Yin, and ELICEIRI K W. Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning[C]. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, USA, 2021: 14313–14323. doi: 10.1109/CVPR46437.2021.01409. [18] CHEN Y C and LU C S. RankMix: Data augmentation for weakly supervised learning of classifying whole slide images with diverse sizes and imbalanced categories[C]. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, Canada, 2023: 23936–23945. doi: 10.1109/CVPR52729.2023.02292. [19] LIU Pei, JI Luping, ZHANG Xinyu, et al. Pseudo-bag mixup augmentation for multiple instance learning-based whole slide image classification[J]. IEEE Transactions on Medical Imaging, 2024, 43(5): 1841–1852. doi: 10.1109/TMI.2024.3351213. [20] YANG Jiawei, CHEN Hanbo, ZHAO Yu, et al. ReMix: A general and efficient framework for multiple instance learning based whole slide image classification[C]. Proceedings of the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, Singapore, 2022: 35–45. doi: 10.1007/978-3-031-16434-7_4. [21] BEJNORDI B E, VETA M, VAN DIEST P J, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer[J]. JAMA, 2017, 318(22): 2199–2210. doi: 10.1001/jama.2017.14585. [22] TOMCZAK K, CZERWIŃSKA P, and WIZNEROWICZ M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge[J]. Contemporary Oncology, 2015, 19(1A): A68–A77. doi: 10.5114/wo.2014.47136. [23] ZHOU S K, RUECKERT D, and FICHTINGER G. Handbook of Medical Image Computing and Computer Assisted Intervention[M]. London: Academic Press, 2020: 521–546. [24] LOU Wei, LI Guanbin, WAN Xiang, et al. Multi-modal denoising diffusion pre-training for whole-slide image classification[C]. Proceedings of the 32nd ACM International Conference on Multimedia, Melbourne, Australia, 2024: 10804–10813. doi: 10.1145/3664647.3680882. -





 下载:
下载:


 下载:
下载:
