Tensor-Train Decomposition for Lightweight Liver Tumor Segmentation
-
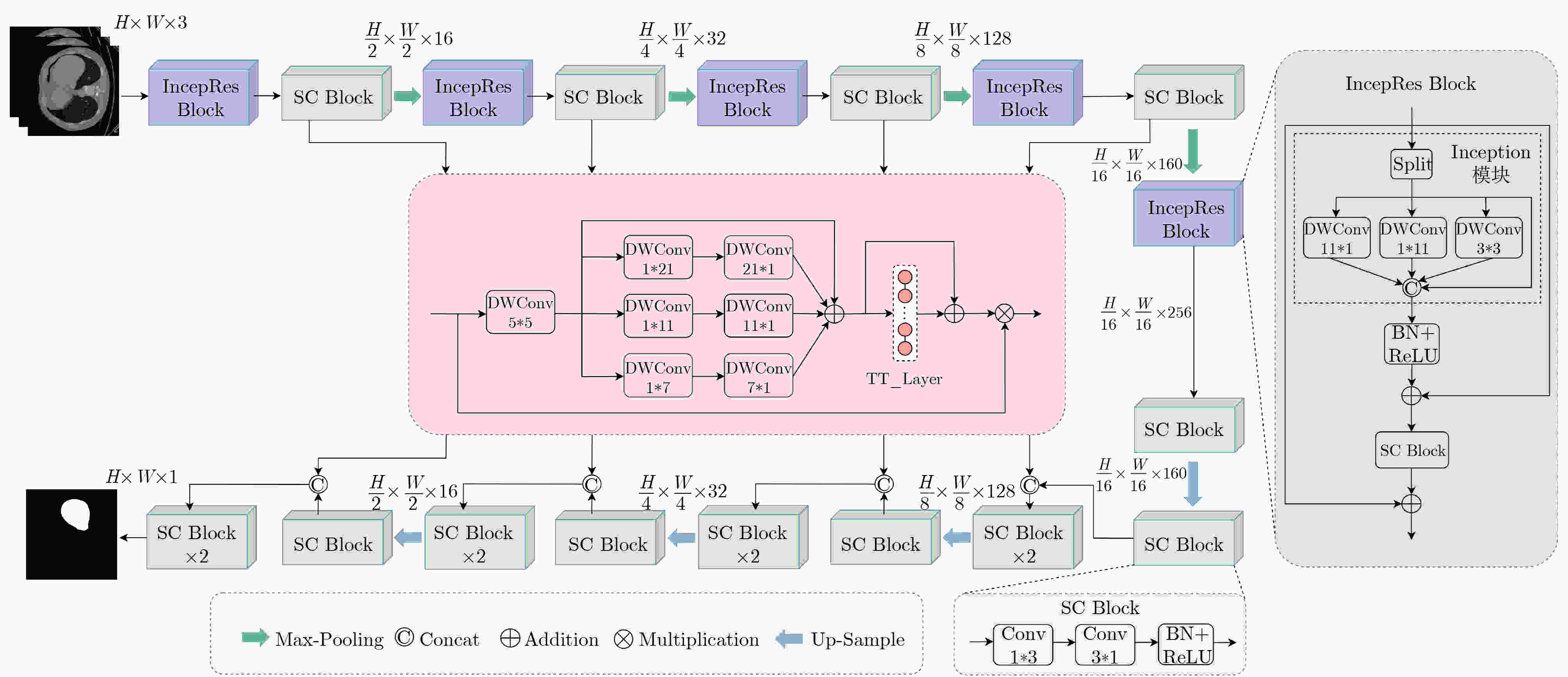
摘要: 针对肝肿瘤分割任务中由于边界复杂性以及肿瘤尺寸较小导致分割结果不准确的问题,该文提出一种高效的轻量化肝肿瘤分割方法。首先,提出一种基于张量列(TT)分解的多尺度卷积注意力(TT-MSCA)模块,通过张量列分解的线性层(TT_Layer)优化多尺度特征融合,提升复杂边界和小尺寸目标的分割准确性;其次,设计一种多分支残差结构的特征提取模块(IncepRes Block),以较小的计算成本提取肝肿瘤图像中的全局上下文信息;最后,解耦标准3*3卷积为两个连续的条形卷积,减少参数量和计算成本。实验结果表明,该方法在LiTS2017和3Dircadb两个公开数据集上,肝脏分割的Dice值分别达到98.54%和97.95%,肿瘤分割的Dice值分别达到94.11%和94.35%。提出方法能够有效解决肝肿瘤边界复杂以及肿瘤目标较小等因素导致的分割结果不准确问题,且能够满足实时部署需求,为肝肿瘤分割提供了一种新的选择。Abstract:
Objective Convolutional Neural Networks (CNNs) have recently achieved notable progress in medical image segmentation. Their conventional convolution operations, however, remain constrained by locality, which reduces their ability to capture global contextual information. Researchers have pursued two main strategies to address this limitation. Hybrid CNN-Transformer architectures use self-attention to model long-range dependencies, and this markedly improves segmentation accuracy. State-space models such as the Mamba series reduce computational cost and retain global modeling capacity, and they also show favorable scalability. Although CNN-Transformer models remain computationally demanding for real-time use, Mamba-based approaches still face challenges such as boundary blur and parameter redundancy when segmenting small targets and low-contrast regions. Lightweight network design has therefore become a research focus. Existing lightweight methods, however, still show limited segmentation accuracy for liver tumor targets with very small sizes and highly complex boundaries. This paper proposes an efficient lightweight method for liver tumor segmentation that aims to meet the combined requirements of high accuracy and real-time performance for small targets with complex boundaries. Methods The proposed method integrates three strategies. A Tensor-Train Multi-Scale Convolutional Attention (TT-MSCA) module is designed to improve segmentation accuracy for small targets and complex boundaries. This module optimizes multi-scale feature fusion through a TT_Layer and employs tensor decomposition to integrate feature information across scales, which supports more accurate identification and segmentation of tumor regions in challenging images. A feature extraction module with a multi-branch residual structure, termed the IncepRes Block, strengthens the model’s capacity to capture global contextual information. Its parallel multi-branch design processes features at several scales and enriches feature representation at a relatively low computational cost. All standard 3*3 convolutions are then decoupled into two consecutive strip convolutions. This reduces the number of parameters and computational cost although the feature extraction capacity is preserved. The combination of these modules allows the method to improve segmentation accuracy and maintain high efficiency, and it demonstrates strong performance for small targets and blurry boundary regions. Results and Discussions Experiments on the LiTS2017 and 3Dircadb datasets show that the proposed method reaches Dice coefficients of 98.54% and 97.95% for liver segmentation, and 94.11% and 94.35% for tumor segmentation. Ablation studies show that the TT-MSCA module and the IncepRes Block improve segmentation performance with only a modest computational cost, and the SC Block reduces computational cost while accuracy is preserved ( Table 2 ). When the TT-MSCA module is inserted into the reduced U-Net on the LiTS2017 dataset, the tumor Dice and IoU reach 93.73% and 83.60%. These values are second only to the final model. On the 3Dircadb dataset, adding the SC Block after TT-MSCA produces a slight accuracy decrease but reduces GFLOPs by a factor of 4.15. Compared with the original U-Net, the present method improves liver IoU by 3.35% and tumor IoU by 5.89%. The TT-MSCA module also consistently exceeds the baseline MSCA module. It increases liver and tumor IoU by 2.59% and 1.95% on LiTS2017, and by 2.03% and 3.13% on 3Dircadb (Table 5 ). These results show that the TT_Layer strengthens global context perception and fine-detail representation through multi-scale feature fusion. The proposed network contains 0.79 M parameters and 1.43 GFLOPs, which represents a 74.9% reduction in parameters compared with CMUNeXt (3.15 M). Real-time performance evaluation records 156.62 fps, more than three times the 50.23 fps of the vanilla U-Net (Table 6 ). Although accuracy decreases slightly in a few isolated metrics, the overall accuracy-compression balance is improved, and the method demonstrates strong practical value for lightweight liver tumor segmentation.Conclusions This paper proposes an efficient liver tumor segmentation method that improves segmentation accuracy and meets real-time requirements. The TT-MSCA module enhances recognition of small targets and complex boundaries through the integration of spatial and channel attention. The IncepRes Block strengthens the network’s perception of liver tumors of different sizes. The decoupling of standard 3*3 convolutions into two consecutive strip convolutions reduces the parameter count and computational cost while preserving feature extraction capacity. Experimental evidence shows that the method reduces errors caused by complex boundaries and small tumor sizes and can satisfy real-time deployment needs. It offers a practical technical option for liver tumor segmentation. The method requires many training iterations to reach optimal data fitting, and future work will address improvements in convergence speed. -
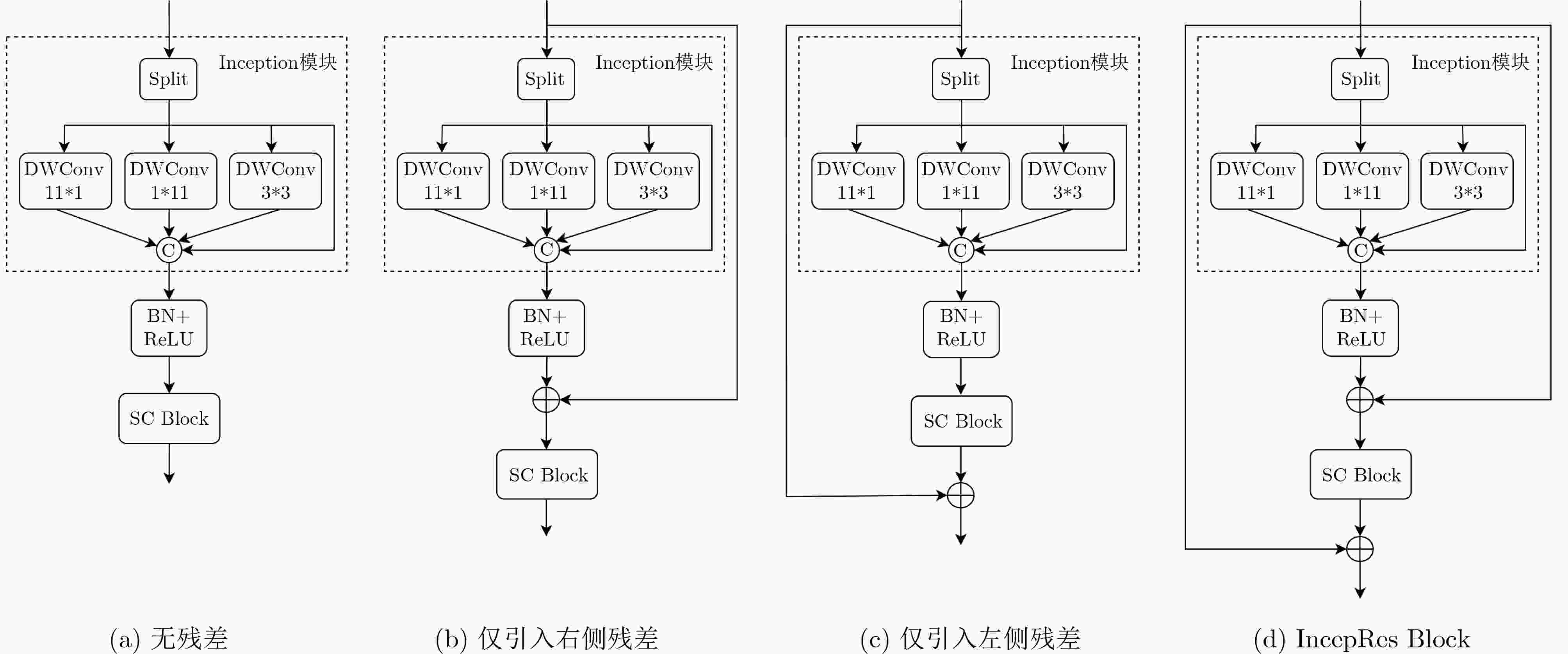
表 1 IncepRes Block内部消融实验结果(%)
各子图编号 LiTS2017 3Dircadb 肝脏 肿瘤 肝脏 肿瘤 Dice IoU Dice IoU Dice IoU Dice IoU (a) 97.35 95.03 92.86 81.95 96.32 92.47 92.48 74.72 (b) 98.12 95.92 93.57 83.65 97.65 93.35 93.46 77.16 (c) 98.01 95.87 93.04 82.13 97.74 93.37 92.85 75.38 (d) 98.54 96.82 94.11 83.91 97.95 94.13 94.35 77.64 注:加粗数值表示最优值,下划线数值表示次优值。 表 2 LiTS2017和3Dircadb数据集上的消融实验结果
Reduced
U-NetTT-
MSCAIncepRes
BlockSC
BlockParams
(M)↓GFLOPs↓ fps↑ LiTS2017(%) 3Dircadb(%) 肝脏 肿瘤 肝脏 肿瘤 Dice IoU Dice IoU Dice IoU Dice IoU - - - - 31.04 54.78 50.23 96.07 94.35 92.28 82.23 96.56 90.78 92.45 71.75 √ - - - 1.91 5.12 172.49 96.12 94.37 92.18 82.15 96.45 90.68 92.33 71.68 √ √ - - 2.21 5.35 149.56 98.35 96.43 93.73 83.60 97.84 93.66 93.28 74.81 √ - √ - 2.03 5.25 178.84 98.47 95.84 93.65 82.34 98.07 93.59 93.27 73.31 √ - - √ 0.37 1.06 282.82 96.02 94.25 92.13 82.11 96.21 90.64 92.26 71.62 √ √ - √ 0.67 1.29 177.92 98.25 96.23 93.57 83.33 97.79 93.64 93.14 74.68 √ - √ √ 0.49 1.19 228.35 98.27 95.04 93.49 82.75 97.90 93.60 92.76 73.49 √ √ √ √ 0.79 1.43 156.62 98.54 96.82 94.11 83.91 97.95 94.13 94.35 77.64 注:加粗数值表示最优值,下划线数值表示次优值。Reduced U-Net(C1=16, C2=32, C3=128, C4=160, C5=256)由原始U-Net(C1=64, C2=128, C3=256, C4=512, C5= 1024 )减少通道数得到。表 3 不同损失函数的性能对比实验结果(%)
损失函数 LiTS2017 3Dircadb 肝脏 肿瘤 肝脏 肿瘤 Dice IoU Dice IoU Dice IoU Dice IoU Dice[25] 97.72 92.48 92.32 81.45 97.13 92.12 90.79 70.72 BCE[26] 98.26 93.57 93.84 79.49 97.87 90.68 91.37 66.03 0.5BCE+Dice[15] 98.56 95.44 93.27 82.12 97.98 94.01 92.35 76.14 Focal[28] 97.79 91.52 91.60 73.58 97.23 88.64 89.66 62.59 Tversky[29] 98.29 93.20 93.85 81.97 98.09 92.37 92.63 71.35 FocalTversky[27] 98.54 96.82 94.11 83.91 97.95 94.13 94.35 77.64 注:加粗数值表示最优值,下划线数值表示次优值。 表 4 FocalTversky损失函数不同权值对比实验结果(%)
编号 $\alpha $ $\beta $ $\gamma $ LiTS2017 3Dircadb 肝脏 肿瘤 肝脏 肿瘤 Dice IoU Dice IoU Dice IoU Dice IoU 1 0.4 0.6 1 97.85 92.51 92.47 81.62 97.58 92.35 91.07 70.75 2 0.5 0.5 1 97.72 92.48 92.32 81.45 97.13 92.12 90.79 70.72 3 0.6 0.4 1 97.96 92.38 92.34 81.23 97.90 91.93 91.92 70.31 4 0.7 0.3 1 98.29 93.20 93.85 81.97 98.09 92.37 92.63 71.35 5 0.8 0.2 1 97.43 91.74 93.62 81.74 96.94 88.41 92.50 71.19 6 0.7 0.3 0.5 98.03 93.12 93.97 82.87 97.84 92.21 93.12 73.77 7 0.7 0.3 0.75 98.54 96.82 94.11 83.91 97.95 94.13 94.35 77.64 8 0.7 0.3 1.25 98.11 92.17 93.43 82.73 97.56 91.01 92.84 73.78 注:加粗数值表示最优值,下划线数值表示次优值。 表 5 不同注意力机制的性能对比实验结果(%)
注意力机制 LiTS2017 3Dircadb 肝脏 肿瘤 肝脏 肿瘤 Dice IoU Dice IoU Dice IoU Dice IoU SE[30] 98.24 93.83 93.79 81.94 97.97 92.47 93.20 74.61 CBAM[31] 97.93 93.01 93.87 81.74 97.85 91.80 93.48 74.58 ECA[32] 97.84 91.43 93.76 81.99 97.99 92.55 93.40 74.72 MSCA[16] 98.29 94.23 93.84 81.96 97.74 92.10 93.78 74.51 TT-MSCA 98.54 96.82 94.11 83.91 97.95 94.13 94.35 77.64 注:加粗数值表示最优值,下划线数值表示次优值。 表 6 LiTS2017数据集上不同方法的性能对比
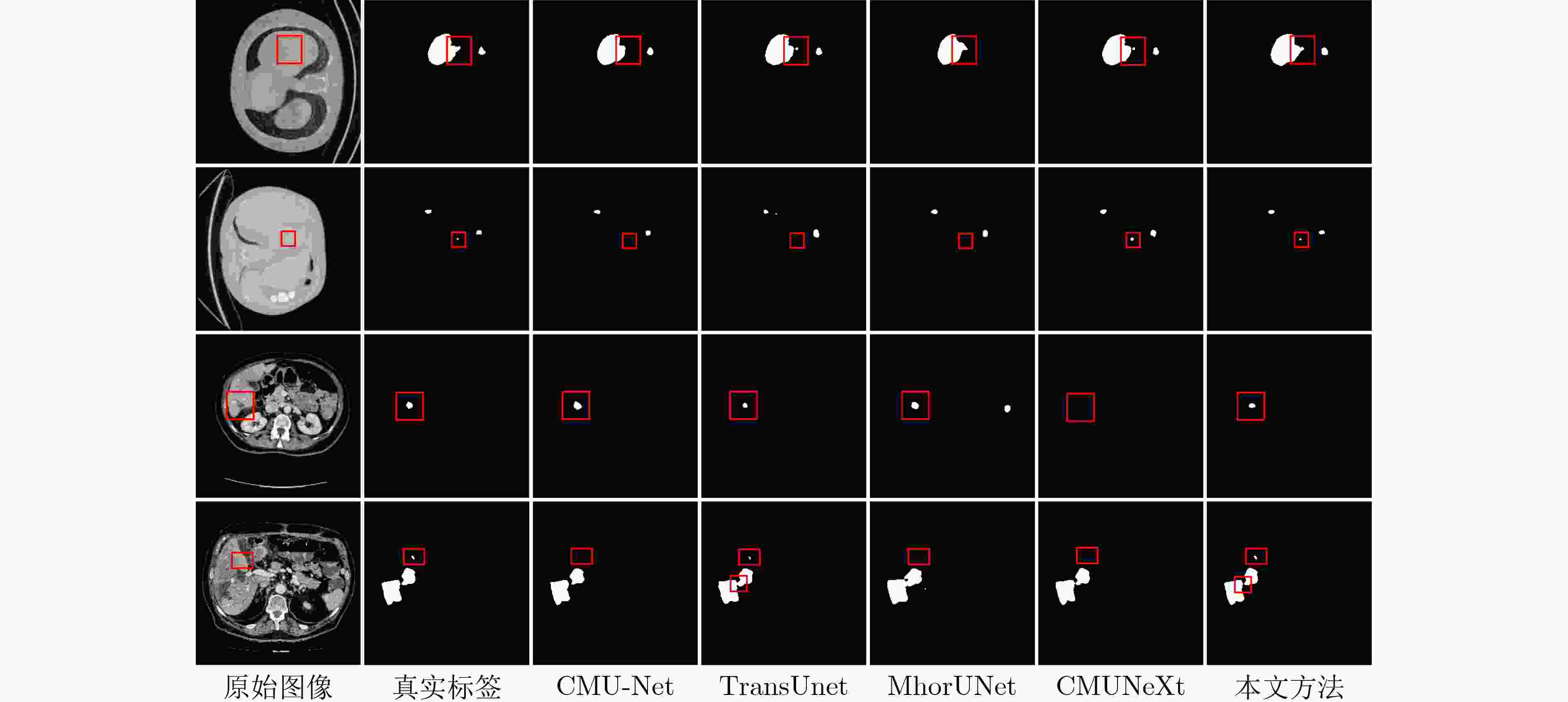
类别 方法 Params(M)↓ GFLOPs↓ fps↑ 肝脏 肿瘤 Dice(%) IoU(%) Dice(%) IoU(%) CNN U-Net[1] 31.04 54.78 50.23 96.07 94.35 92.28 82.23 U-Net3+[2] 26.97 199.74 28.53 96.44 87.19 92.81 79.61 CMU-Net[3] 49.93 91.25 41.48 98.29 96.07 93.28 81.47 CSCA U-Net[4] 35.27 13.74 62.96 98.41 94.73 93.42 83.80 CNN-
TransformerTransUnet[5] 105.32 38.52 42.52 98.20 96.70 93.35 82.16 SwinUnet[6] 27.14 5.91 132.58 97.86 91.73 92.19 80.25 SSTrans-Net[7] 29.64 6.02 125.73 98.12 95.82 93.09 80.40 轻量化 MhorUNet[12] 3.53 0.58 13.72 97.94 95.50 92.91 80.04 ConvUNeXt[14] 3.50 7.25 146.72 97.55 93.98 93.16 82.09 CMUNeXt[15] 3.15 7.42 151.53 98.36 96.42 93.57 83.17 本文方法 0.79 1.43 156.62 98.54 96.82 94.11 83.91 注:加粗数值表示最优值,下划线数值表示次优值。 表 7 3Dircadb数据集上不同方法的性能对比
类别 方法 Params(M)↓ GFLOPs↓ fps↑ 肝脏 肿瘤 Dice(%) IoU(%) Dice(%) IoU(%) CNN U-Net[1] 31.04 54.78 50.23 96.56 90.78 92.45 71.75 U-Net3+[2] 26.97 199.74 28.53 96.89 89.41 93.29 75.32 CMU-Net[3] 49.93 91.25 41.48 97.64 92.23 94.27 79.09 CSCA U-Net[4] 35.27 13.74 62.96 97.93 93.72 93.92 78.46 CNN-
TransformerTransUnet[5] 105.32 38.52 42.52 97.89 93.46 93.29 74.42 SwinUnet[6] 27.14 5.91 132.58 96.42 89.93 91.73 74.61 SSTrans-Net[7] 29.64 6.02 125.73 97.45 91.27 92.94 74.98 轻量化 MhorUNet[12] 3.53 0.58 13.72 96.25 87.90 90.63 73.64 ConvUNeXt[14] 3.50 7.25 146.72 96.50 87.43 91.92 72.10 CMUNeXt[15] 3.15 7.42 151.53 97.79 91.39 93.58 76.28 本文方法 0.79 1.43 156.62 97.95 94.13 94.35 77.64 注:加粗数值表示最优值,下划线数值表示次优值。 -
[1] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. The 18th International Conference on Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [2] HUANG Huimin, LIN Lanfen, TONG Ruofeng, et al. UNet 3+: A full-scale connected unet for medical image segmentation[C]. The ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing, Barcelona, Spain, 2020: 1055–1059. doi: 10.1109/ICASSP40776.2020.9053405. [3] TANG Fenghe, WANG Lingtao, NING Chunping, et al. CMU-Net: A strong convmixer-based medical ultrasound image segmentation network[C]. 2023 IEEE 20th International Symposium on Biomedical Imaging, Cartagena, Colombia, 2023: 1–5. doi: 10.1109/ISBI53787.2023.10230609. [4] SHU Xin, WANG Jiashu, ZHANG Aoping, et al. CSCA U-Net: A channel and space compound attention CNN for medical image segmentation[J]. Artificial Intelligence in Medicine, 2024, 150: 102800. doi: 10.1016/j.artmed.2024.102800. [5] CHEN Jieneng, LU Yongyi, YU Qihang, et al. TransUNet: Transformers make strong encoders for medical image segmentation[EB/OL]. https://arxiv.org/abs/2102.04306, 2021. [6] CAO Hu, WANG Yueyue, CHEN J, et al. Swin-Unet: Unet-like pure transformer for medical image segmentation[M]. KARLINSKY L, MICHAELI T, and NISHINO K. Computer Vision– ECCV 2022 Workshops. Cham: Springer, 2023: 205–218. doi: 10.1007/978-3-031-25066-8_9. [7] FU Liyao, CHEN Yunzhu, JI Wei, et al. SSTrans-Net: Smart swin transformer network for medical image segmentation[J]. Biomedical Signal Processing and Control, 2024, 91: 106071. doi: 10.1016/j.bspc.2024.106071. [8] XING Zhaohu, YE Tian, YANG Yijun, et al. SegMamba: Long-range sequential modeling mamba for 3D medical image segmentation[C]. The 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 578–588. doi: 10.1007/978-3-031-72111-3_54. [9] DUTTA T K, MAJHI S, NAYAK D R, et al. SAM-Mamba: Mamba guided SAM architecture for generalized zero-shot polyp segmentation[C]. 2025 IEEE/CVF Winter Conference on Applications of Computer Vision, Tucson, USA, 2025: 4655–4664. doi: 10.1109/WACV61041.2025.00457. [10] KUI Xiaoyan, JIANG Shen, LI Qinsong, et al. Gl-MambaNet: A global-local hybrid Mamba network for medical image segmentation[J]. Neurocomputing, 2025, 626: 129580. doi: 10.1016/j.neucom.2025.129580. [11] VALANARASU J M J and PATEL V M. UNeXt: MLP-based rapid medical image segmentation network[C]. The 25th International Conference on Medical Image Computing and Computer Assisted Intervention, Singapore, Singapore, 2022: 23–33. doi: 10.1007/978-3-031-16443-9_3. [12] WU Renkai, LIANG Pengchen, HUANG Xuan, et al. MHorUNet: High-order spatial interaction UNet for skin lesion segmentation[J]. Biomedical Signal Processing and Control, 2024, 88: 105517. doi: 10.1016/j.bspc.2023.105517. [13] TROCKMAN A and KOLTER J Z. Patches are all you need?[EB/OL]. https://arxiv.org/abs/2201.09792, 2022. [14] HAN Zhimeng, JIAN Muwei, and WANG Gaige. ConvUNeXt: An efficient convolution neural network for medical image segmentation[J]. Knowledge-Based Systems, 2022, 253: 109512. doi: 10.1016/j.knosys.2022.109512. [15] TANG Fenghe, DING Jianrui, QUAN Quan, et al. CMUNEXT: An efficient medical image segmentation network based on large kernel and skip fusion[C]. 2024 IEEE International Symposium on Biomedical Imaging, Athens, Greece, 2024: 1–5. doi: 10.1109/ISBI56570.2024.10635609. [16] GUO Menghao, LU Chengze, HOU Qibin, et al. SegNeXt: Rethinking convolutional attention design for semantic segmentation[C]. The 36th International Conference on Neural Information Processing Systems, New Orleans, USA, 2022: 84. doi: 10.5555/3600270.3600354. [17] GARIPOV T, PODOPRIKHIN D, NOVIKOV A, et al. Ultimate tensorization: Compressing convolutional and FC layers alike[EB/OL]. https://arxiv.org/abs/1611.03214, 2016. [18] YAN Jiale, ANDO K, YU J, et al. TT-MLP: Tensor train decomposition on deep MLPs[J]. IEEE Access, 2023, 11: 10398–10411. doi: 10.1109/ACCESS.2023.3240784. [19] YU Weihao, ZHOU Pan, YAN Shuicheng, et al. InceptionNeXt: When inception meets convnext[C]. 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, USA, 2024: 5672–5683. doi: 10.1109/CVPR52733.2024.00542. [20] SZEGEDY C, VANHOUCKE V, IOFFE S, et al. Rethinking the inception architecture for computer vision[C]. 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 2818–2826. doi: 10.1109/CVPR.2016.308. [21] LIAO Juan, CHEN Minhui, ZHANG Kai, et al. SC-Net: A new strip convolutional network model for rice seedling and weed segmentation in paddy field[J]. Computers and Electronics in Agriculture, 2024, 220: 108862. doi: 10.1016/j.compag.2024.108862. [22] MA Yuliang, WU Liping, GAO Yunyuan, et al. ULFAC-Net: Ultra-lightweight fully asymmetric convolutional network for skin lesion segmentation[J]. IEEE Journal of Biomedical and Health Informatics, 2023, 27(6): 2886–2897. doi: 10.1109/jbhi.2023.3259802. [23] BILIC P, CHRIST P, LI H B, et al. The liver tumor segmentation benchmark (LiTS)[J]. Medical Image Analysis, 2023, 84: 102680. doi: 10.1016/j.media.2022.102680. [24] CHRIST P F, ELSHAER M E A, ETTLINGER F, et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields[C].The 19th International Conference on Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, Athens, Greece, 2016: 415–423. doi: 10.1007/978-3-319-46723-8_48. [25] JIANG Huiyan, DIAO Zhaoshuo, SHI Tianyu, et al. A review of deep learning-based multiple-lesion recognition from medical images: Classification, detection and segmentation[J]. Computers in Biology and Medicine, 2023, 157: 106726. doi: 10.1016/j.compbiomed.2023.106726. [26] KHARE N, THAKUR P S, KHANNA P, et al. Analysis of loss functions for image reconstruction using convolutional autoencoder[C]. The 6th International Conference on Computer Vision and Image Processing, Rupnagar, India, 2021: 338–349. doi: 10.1007/978-3-031-11349-9_30. [27] ABRAHAM N and KHAN N M. A novel focal tversky loss function with improved attention U-Net for lesion segmentation[C]. 2019 IEEE 16th International Symposium on Biomedical Imaging, Venice, Italy, 2019: 683–687. doi: 10.1109/ISBI.2019.8759329. [28] LIN T Y, GOYAL P, GIRSHICK R, et al. Focal loss for dense object detection[C]. 2007 IEEE International Conference on Computer Vision, Venice, Italy, 2017: 2999–3007. doi: 10.1109/ICCV.2017.324. [29] MONTAZEROLGHAEM M, SUN Yu, SASSO G, et al. U-Net architecture for prostate segmentation: The impact of loss function on system performance[J]. Bioengineering, 2023, 10(4): 412. doi: 10.3390/bioengineering10040412. [30] HU Jie, SHEN Li, and SUN Gang. Squeeze-and-excitation networks[C]. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 7132–7141. doi: 10.1109/CVPR.2018.00745. [31] WOO S, PARK J, LEE J Y, et al. CBAM: Convolutional block attention module[C]. The 15th European Conference on Computer Vision, Munich, Germany, 2018: 3–19. doi: 10.1007/978-3-030-01234-2_1. [32] WANG Qilong, WU Banggu, ZHU Pengfei, et al. ECA-Net: Efficient channel attention for deep convolutional neural networks[C]. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, USA, 2020: 11531–11539. doi: 10.1109/CVPR42600.2020.01155. -





 下载:
下载:


 下载:
下载:
