Global Perception and Sparse Feature Associate Image-level Weakly Supervised Pathological Image Segmentation
-
摘要: 弱监督语义分割方法可以节省大量的人工标注成本,在病理全切片图像(WSI)的分析中有着广泛应用。针对弱监督多实例学习(MIL)方法在病理图像分析中存在的像素实例相互独立缺乏依赖关系,分割结果局部不一致和图像级标签监督信息不充分的问题,该文提出一种全局感知与稀疏特征关联图像级弱监督的端到端多实例学习方法(DASMob-MIL)。首先,为克服像素实例之间的独立性,使用局部感知网络提取特征以建立局部像素依赖,并级联交叉注意力模块构建全局信息感知分支(GIPB)以建立全局像素依赖关系。其次,引入像素自适应细化模块(PAR),通过多尺度邻域局部稀疏特征之间的相似性构建亲和核,解决了弱监督语义分割结果局部不一致的问题。最后,设计深度关联监督模块(DAS),通过对多阶段特征图生成的分割图进行加权融合,并使用权重因子关联损失函数以优化训练过程,以降低弱监督图像级标签监督信息不充分的影响。DASMob-MIL模型在自建的结直肠癌数据集YN-CRC和公共弱监督组织病理学图像数据集LUAD-HistoSeg-BC上与其他模型相比展示出了先进的分割性能,模型权重仅为14 MB,在YN-CRC数据集上F1 Score达到了89.5%,比先进的多层伪监督(MLPS)模型提高了3%。实验结果表明,DASMob-MIL仅使用图像级标签实现了像素级的分割,有效改善了弱监督组织病理学图像的分割性能。Abstract: The weakly supervised semantic segmentation methods have been widely applied in the analysis of Whole Slide Images (WSI), saving a considerable amount of manual annotation costs. Addressing the issues of pixel instance independence, local inconsistency in segmentation results, and insufficient supervision from image-level labels in Multiple-Instance Learning (MIL) methods for pathological image analysis, a novel end-to-end MIL approach named DASMob-MIL is proposed in this paper. Firstly, to overcome the independence among pixel instances, features are extracted using a local perception network to establish local pixel dependencies, while a Global Information Perception Branch (GIPB) is constructed by cascading cross-attention modules to establish global pixel dependencies. Secondly, a Pixel-Adaptive Refinement (PAR) module is introduced to address the problem of local inconsistency in weakly supervised semantic segmentation results by constructing affinity kernels based on the similarity between multi-scale neighborhood local sparse features. Finally, a Deep Association Supervision (DAS) module is designed to optimize the training process by performing weighted fusion on the segmentation maps generated from multi-stage feature maps. Then, employing a weighted factor-associated loss function to mitigate the impact of insufficient supervision from weakly supervised image-level labels. Compared with other models, the DASMob-MIL model demonstrates advanced segmentation performance on the self-built colorectal cancer dataset YN-CRC and the public weakly supervised histopathology image dataset LUAD-HistoSeg-BC, with a model weight of only 14MB and an F1 score of 89.5% on the YN-CRC dataset, which was 3% higher than that of the advanced Multi-Layer Pseudo-Supervision (MLPS) model. Experimental results indicate that DASMob-MIL achieves pixel-level segmentation utilizing only image-level labels, effectively improving the segmentation performance of weakly supervised histopathological images.
-
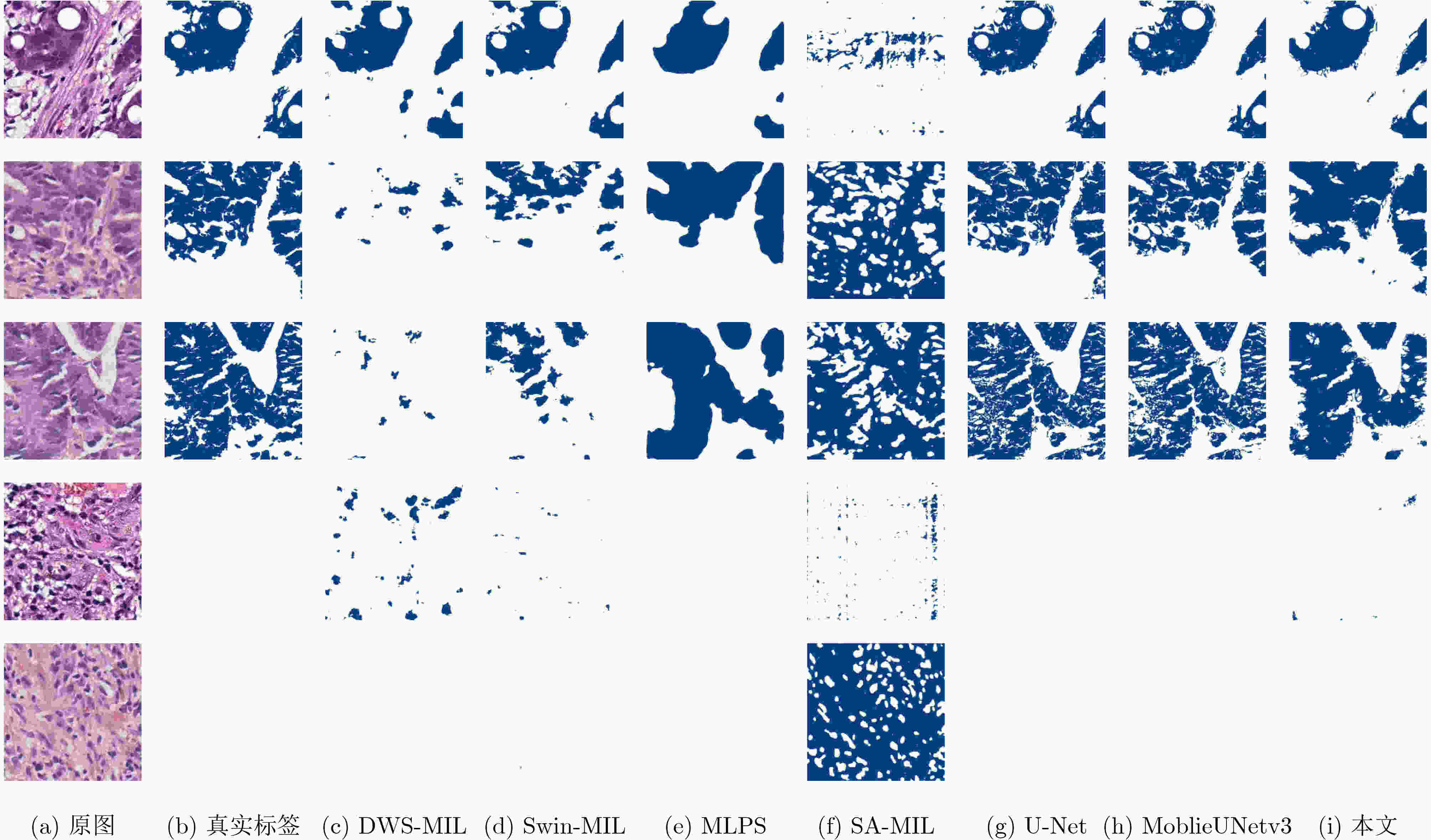
表 1 不同模型在YN-CRC数据集上的分割性能对比
模型 F1 EC (%) F1 NEC (%) F1 Score (%) HD EC Precision (%) Recall (%) 权重 (MB) 推理时间(s) 全监督 U-Net 91.4 99.6 93.0 5.973 95.1 91.4 33.0 0.0112 MobileUNetv3 91.6 99.6 93.1 5.378 95.2 91.6 26.6 0.0056 弱监督 SA-MIL 35.4 87.5 45.3 42.103 61.8 43.0 7.07 0.1218 DWS-MIL 76.7 98.7 80.9 27.690 89.5 82.4 6.65 0.0144 Swin-MIL 82.9 99.6 86.1 18.915 90.3 86.3 105 0.0279 MLPS 83.4 99.8 86.5 41.701 83.8 91.7 453 0.0220 本文(DASMob-MIL) 87.3 99.0 89.5 23.576 86.5 94.6 14.0 0.0712 表 2 不同模型在LUAD-HistoSeg-BC数据集上的分割性能对比
模型 F1 TM (%) F1 NTM (%) F1 Score (%) HD TM Precision (%) Recall (%) 权重(MB) 推理时间(s) 弱监督 MLPS 56.9 99.9 61.8 38.029 76.4 56.7 453 0.0133 SA-MIL 65.9 100 69.8 19.012 78.6 70.8 7.07 0.0268 DWS-MIL 68.5 94.9 71.5 19.578 76.9 75.9 6.65 0.0079 Swin-MIL 71.6 99.4 74.7 19.148 74.5 82.5 105 0.0209 本文(DASMob-MIL) 73.4 98.5 76.3 23.515 73.6 84.6 14.0 0.0378 表 3 不同局部特征提取主干对分割精度的影响
主干 F1 EC(%) F1 NEC(%) F1 Score(%) HD EC Precision(%) Recall(%) 权重(MB) 推理时间(s) VGG-16 59.9 100 67.5 159.929 57.2 98.4 100 0.0624 ResNet50 70.7 99.8 76.2 42.565 74.6 85.8 281 0.0349 EfficientNetv2 73.2 99.6 78.2 78.894 72.0 91.3 212 0.0463 ShuffleNetv2 75.5 99.4 80.0 73.642 75.5 90.0 69.0 0.0185 U-Net 78.2 98.4 82.1 64.231 74.0 95.5 65.9 0.0364 MobileNetv3 80.1 99.4 83.7 26.621 86.2 86.3 13.3 0.0143 表 4 所提出的模块对分割精度的影响
模型 模块 评价指标 GIPB PAR DAS F1 EC (%) F1 NEC (%) F1 Score (%) HD EC Precision (%) Recall (%) 权重(MB) 推理时间(s) 基准 80.1 99.4 83.7 26.621 86.2 86.3 13.3 0.0143 消融1 √ 82.4 99.6 85.7 15.667 86.3 87.3 13.8 0.0150 消融2 √ 83.4 99.5 86.4 22.674 88.4 87.6 13.5 0.0285 消融3 √ 84.5 99.7 87.4 28.712 86.8 90.5 13.3 0.0427 消融4 √ √ 83.8 98.3 86.5 18.664 82.0 93.8 14.0 0.0316 消融5 √ √ 85.4 99.5 88.1 27.261 89.5 89.2 13.5 0.0625 消融6 √ √ 86.0 99.3 88.6 25.358 84.6 95.1 13.9 0.0448 DASMob-MIL √ √ √ 87.3 99.0 89.5 23.576 86.5 94.6 14.0 0.0712 表 5 PAR模块中迭代次数对分割精度的影响
$ T $ F1 EC(%) F1 NEC(%) F1 Score(%) HD EC Precision(%) Recall(%) 推理时间(s) 基准 80.1 99.4 83.7 26.621 86.2 86.3 0.0143 5 80.6 99.8 84.3 38.394 84.5 88.8 0.0341 10 84.5 99.7 87.4 28.712 86.8 90.5 0.0427 15 83.7 99.7 86.7 33.183 86.5 90.7 0.0529 20 79.9 99.4 83.6 41.216 83.2 88.5 0.0640 表 6 不同GIPB配置对分割精度的影响
编码器数 F1 EC(%) F1 NEC(%) F1 Score(%) HD EC Precision(%) Recall(%) 权重(MB) 推理时间(s) 基准 80.1 99.4 83.7 26.621 86.2 86.3 13.3 0.0143 1 80.1 99.8 83.9 16.783 84.9 87.0 13.4 0.0278 2 78.3 99.4 82.3 35.055 82.2 87.0 13.4 0.0265 3 83.4 99.5 86.4 22.674 88.4 87.6 13.5 0.0285 4 79.9 98.9 83.4 30.955 84.0 87.8 14.1 0.0328 5 75.6 99.8 80.2 34.782 84.2 82.5 16.0 0.0346 表 7 DAS结构中不同侧分支权重系数对分割精度的影响
分组 权重系数 F1 EC(%) F1 NEC(%) F1 Score(%) HD EC Precision(%) Recall(%) 推理时间(s) 基准 80.1 99.4 83.7 26.621 86.2 86.3 0.0143 1 [0.15,0.15,0.2,0.5] 82.4 99.6 85.7 15.667 86.3 87.3 0.0150 2 [0.1,0.1,0.3,0.5] 81.2 99.7 84.7 19.489 88.0 86.5 0.0151 3 [0.15,0.15,0.3,0.4] 74.3 99.7 79.1 21.112 75.2 88.8 0.0149 4 [0.2,0.2,0.3,0.3] 80.6 97.9 83.9 21.314 80.3 90.5 0.0152 5 [0.2,0.2,0.25,0.35] 81.2 99.6 84.7 20.356 83.0 90.6 0.0150 -
[1] BRAY F, FERLAY J, SOERJOMATARAM I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2018, 68(6): 394–424. doi: 10.3322/caac.21492. [2] ZIDAN U, GABER M M, and ABDELSAMEA M M. SwinCup: Cascaded swin transformer for histopathological structures segmentation in colorectal cancer[J]. Expert Systems with Applications, 2023, 216: 119452. doi: 10.1016/j.eswa.2022.119452. [3] JIA Zhipeng, HUANG Xingyi, CHANG E I C, et al. Constrained deep weak supervision for histopathology image segmentation[J]. IEEE Transactions on Medical Imaging, 2017, 36(11): 2376–2388. doi: 10.1109/TMI.2017.2724070. [4] CAI Hongmin, YI Weiting, LI Yucheng, et al. A regional multiple instance learning network for whole slide image segmentation[C]. 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, USA, 2022: 922–928. doi: 10.1109/BIBM55620.2022.9995017. [5] LI Kailu, QIAN Ziniu, HAN Yingnan, et al. Weakly supervised histopathology image segmentation with self-attention[J]. Medical Image Analysis, 2023, 86: 102791. doi: 10.1016/j.media.2023.102791. [6] ZHOU Yanzhao, ZHU Yi, YE Qixiang, et al. Weakly supervised instance segmentation using class peak response[C]. The 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 3791–3800. doi: 10.1109/CVPR.2018.00399. [7] ZHONG Lanfeng, WANG Guotai, LIAO Xin, et al. HAMIL: High-resolution activation maps and interleaved learning for weakly supervised segmentation of histopathological images[J]. IEEE Transactions on Medical Imaging, 2023, 42(10): 2912–2923. doi: 10.1109/TMI.2023.3269798. [8] HAN Chu, LIN Jiatai, MAI Jinhai, et al. Multi-layer pseudo-supervision for histopathology tissue semantic segmentation using patch-level classification labels[J]. Medical Image Analysis, 2022, 80: 102487. doi: 10.1016/j.media.2022.102487. [9] DIETTERICH T G, LATHROP R H, and LOZANO-PÉREZ T. Solving the multiple instance problem with axis-parallel rectangles[J]. Artificial Intelligence, 1997, 89(1/2): 31–71. doi: 10.1016/S0004-3702(96)00034-3. [10] XU Gang, SONG Zhigang, SUN Zhuo, et al. CAMEL: A weakly supervised learning framework for histopathology image segmentation[C]. The 2019 IEEE/CVF International Conference on Computer Vision, Seoul, Korea (South), 2019: 10681–10690. doi: 10.1109/ICCV.2019.01078. [11] 徐金东, 赵甜雨, 冯国政, 等. 基于上下文模糊C均值聚类的图像分割算法[J]. 电子与信息学报, 2021, 43(7): 2079–2086. doi: 10.11999/JEIT200263.XU Jindong, ZHAO Tianyu, FENG Guozheng, et al. Image segmentation algorithm based on context fuzzy C-means clustering[J]. Journal of Electronics & Information Technology, 2021, 43(7): 2079–2086. doi: 10.11999/JEIT200263. [12] 杭昊, 黄影平, 张栩瑞, 等. 面向道路场景语义分割的移动窗口变换神经网络设计[J]. 光电工程, 2024, 51(1): 230304. doi: 10.12086/oee.2024.230304.HANG Hao, HUANG Yingping, ZHANG Xurui, et al. Design of Swin Transformer for semantic segmentation of road scenes[J]. Opto-Electronic Engineering, 2024, 51(1): 230304. doi: 10.12086/oee.2024.230304. [13] QIAN Ziniu, LI Kailu, LAI Maode, et al. Transformer based multiple instance learning for weakly supervised histopathology image segmentation[C]. The 25th International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 2022: 160–170. doi: 10.1007/978-3-031-16434-7_16. [14] HUANG Zilong, WANG Xinggang, HUANG Lichao, et al. CCNet: Criss-cross attention for semantic segmentation [C]. The IEEE/CVF International Conference on Computer Vision, Seoul, Korea (South), 2019: 603–612. doi: 10.1109/ICCV.2019.00069. [15] RU Lixiang, ZHAN Yibing, YU Baosheng, et al. Learning affinity from attention: End-to-end weakly-supervised semantic segmentation with transformers[C]. The 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, USA, 2022: 16825–16834. doi: 10.1109/CVPR52688.2022.01634. [16] XIE Yuhan, ZHANG Zhiyong, CHEN Shaolong, et al. Detect, Grow, Seg: A weakly supervision method for medical image segmentation based on bounding box[J]. Biomedical Signal Processing and Control, 2023, 86: 105158. doi: 10.1016/j.bspc.2023.105158. [17] KWEON H, YOON S H, KIM H, et al. Unlocking the potential of ordinary classifier: Class-specific adversarial erasing framework for weakly supervised semantic segmentation[C]. The 2021 IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 6974–6983. doi: 10.1109/ICCV48922.2021.00691. [18] HOWARD A, SANDLER M, CHEN Bo, et al. Searching for MobileNetV3[C]. The 2019 IEEE/CVF International Conference on Computer Vision, Seoul, Korea (South), 2019: 1314–1324. doi: 10.1109/ICCV.2019.00140. [19] VIOLA P, PLATT J C, and ZHANG Cha. Multiple instance boosting for object detection[J]. Proceedings of the 18th International Conference on Neural Information Processing Systems, Vancouver, Canada, 2005: 1417–1424. [20] RONNEBERGER O, FISCHER P, and BROX T. U-net: Convolutional networks for biomedical image segmentation[C]. The 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [21] SIMONYAN K and ZISSERMAN A. Very deep convolutional networks for large-scale image recognition[C]. The 3rd International Conference on Learning Representations, San Diego, USA, 2015. [22] HE Kaiming, ZHANG Xiangyu, REN Shaoqing, et al. Deep residual learning for image recognition[C]. The 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 770–778. doi: 10.1109/CVPR.2016.90. [23] MA Ningning, ZHANG Xiangyu, ZHENG Haitao, et al. ShuffleNet v2: Practical guidelines for efficient CNN architecture design[C]. The 15th European Conference on Computer Vision, Munich, Germany, 2018: 122–138. doi: 10.1007/978-3-030-01264-9_8. [24] TAN Mingxing and LE Q V. EfficientNetV2: Smaller models and faster training[C]. The 38th International Conference on Machine Learning, 2021: 10096–10106. -






 下载:
下载:





 下载:
下载:
