A Fusion-based Approach for Cervical Cell Classification Incorporating Personalized Relationships and Background Information
-
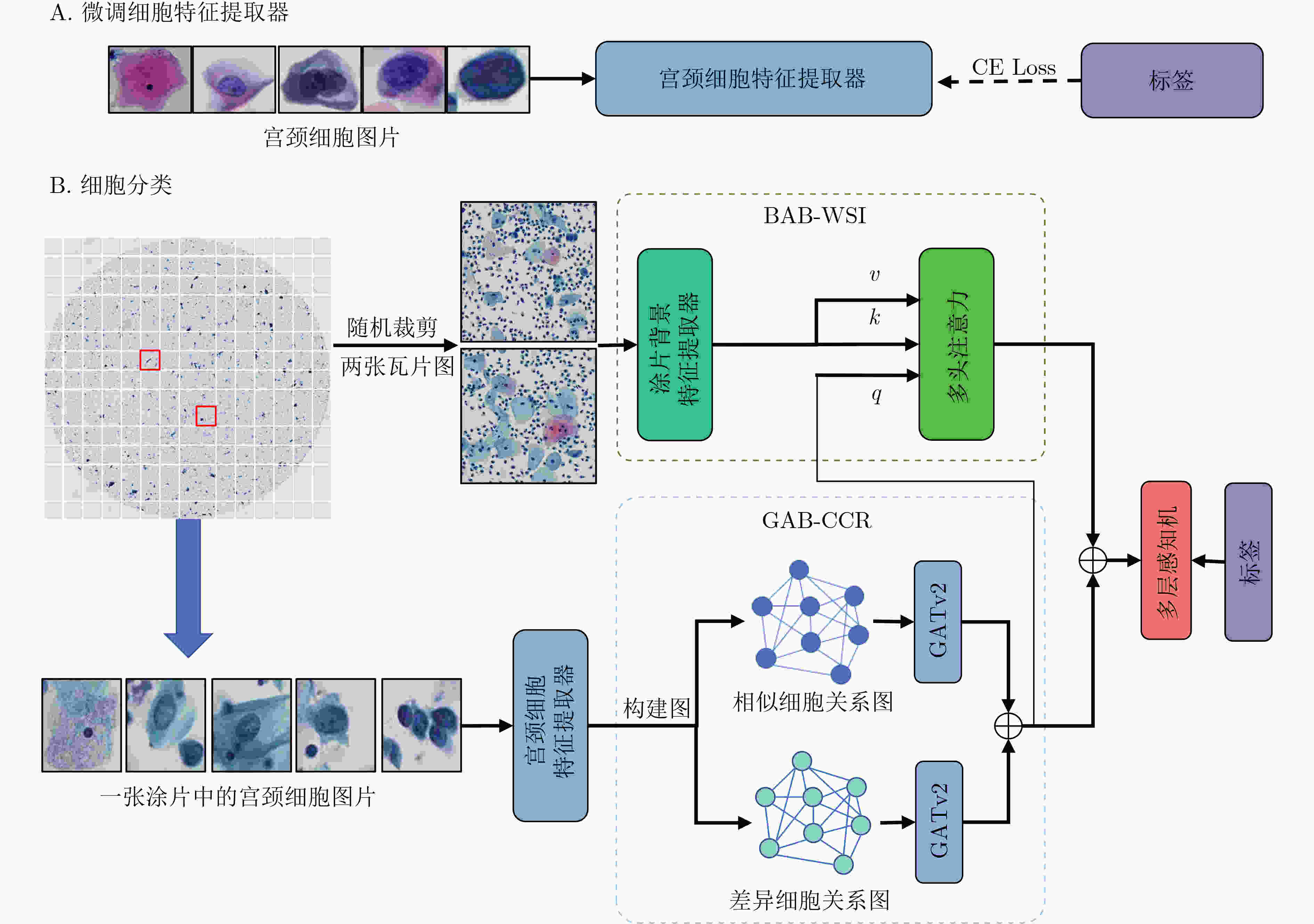
摘要: 宫颈细胞分类在宫颈癌辅助诊断中发挥着重要的作用。然而,现有的宫颈细胞分类方法未充分考虑细胞关系和背景信息,也没有模拟病理医生的诊断方式,导致分类性能较低。因此,该文提出了一种融合细胞关系和背景信息的宫颈细胞分类方法,由基于细胞关系的图注意力分支(GAB-CCR)和背景信息注意力分支(BAB-WSI)组成。GAB-CCR采用细胞特征间的余弦相似度,首先构建相似和差异细胞关系图,并利用GATv2增强模型对细胞关系建模。BAB-WSI使用多头注意力模块捕捉涂片背景上的关键信息并反映不同区域的重要性。最后,将增强后的细胞特征和背景特征融合,提升了网络的分类性能。实验表明,相比于基线模型Swin Transformer-L,所提方法在准确率、敏感度、特异性和F1-Score分别提高了15.9%, 30.32%, 8.11%和31.62%。Abstract: Cervical cell classification plays a crucial role in assisting the diagnosis of cervical cancer. However, existing methods for cervical cell classification do not enough consider relationships among cells and background information, and fail to effectively simulate the diagnostic approach of pathology doctors. As a result, their classification performance is limited. In this study, a novel approach that integrates cell relationships and background information for cervical cell classification is proposed. The proposed method consists of a Graph Attention Branching for Cell-Cell Relationships (GAB-CCR) and a Background Attention Branching for Whole Slide Images (BAB-WSI). GAB-CCR utilizes cosine similarity of cell features to construct preliminary graphs representing similar and distinct cell relationships. Additionally, GAB-CCR enhances the ability of models in modeling cell relationships through GATv2. BAB-WSI employs multi-head attention to effectively capture crucial information on the slide background and reflect the importance of different regions. Finally, the enhanced cell and background features are fused to improve the classification performance of the network. Experimental results demonstrate that the proposed method achieves significant improvements over the baseline model, Swin Transformer-L, with improvement in accuracy, sensitivity, specificity, and F1-Score by 15.9%, 30.32%, 8.11%, and 31.62% respectively.
-
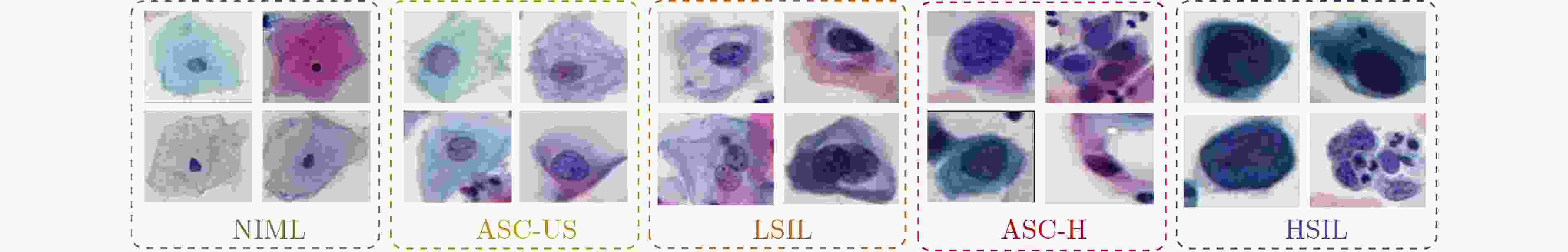
表 1 不同类别的宫颈细胞数量分布
细胞类别 细胞数量 类型 NIML 14516 正常 ASCUS 15897 异常 LSIL 6631 异常 ASCH 2238 异常 HSIL 2229 异常 总计 41511 – 表 2 对比实验结果(%)
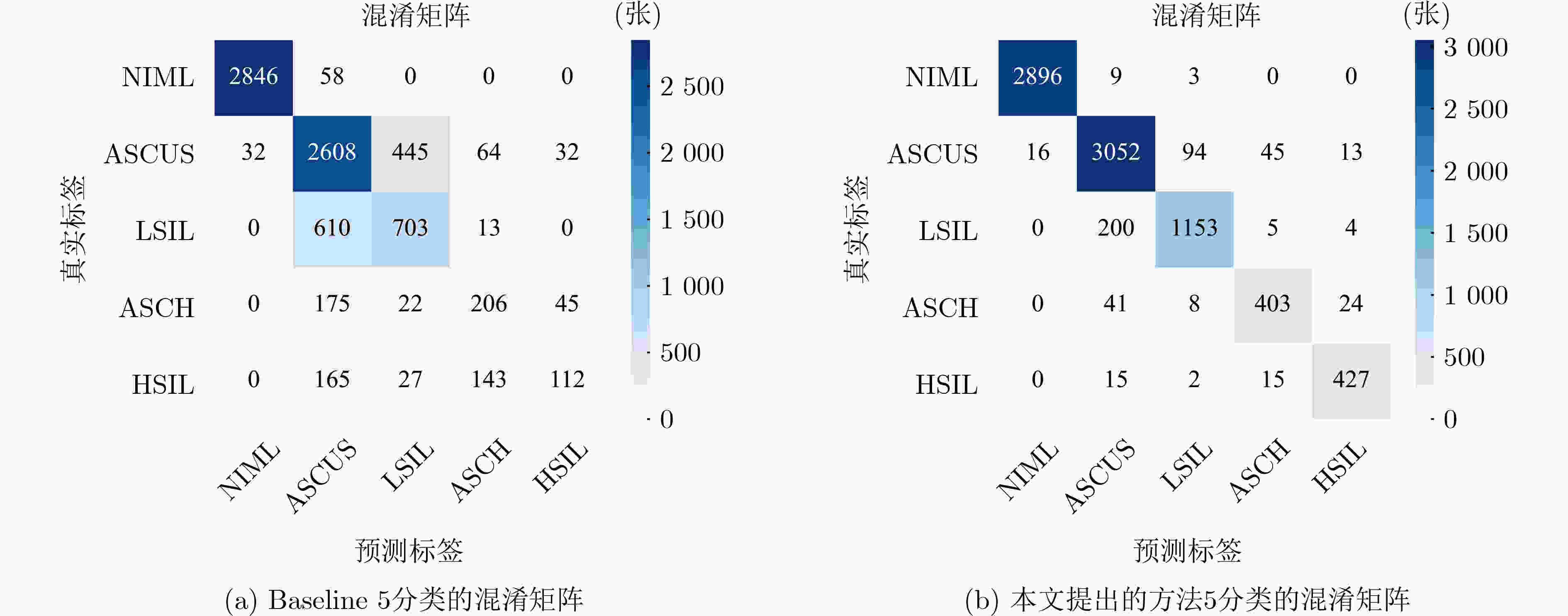
方法 准确率 敏感度 特异性 F1-Score Resnet152 78.62 63.25 93.95 65.15 DenseNet121 78.96 63.73 90.93 63.43 Inception v3 78.35 61.45 90.36 58.87 Efficientnetv2-L 80.36 67.95 94.63 68.57 VOLO-D2/224 79.03 64 91 58.87 VIT-B 77.35 59.33 89.83 58.87 Xcit-s24 76.19 61.88 90.47 58.87 Swin Transformer-L 78.23 61.02 90.25 58.87 Mixer-B/16 76.79 59.83 89.95 58.87 DeepCervix 76.71 60.22 93.51 61.76 Basak等人[19] 70.83 42.46 90.87 42.41 Manna等人[17] 72.97 53.13 92.10 55.49 Shi等人[15] 88.48 88.51 97.01 88.33 Ou等人[40] 89.06 89.25 97.15 89.13 本文方法 94.13 91.34 98.36 91.67 表 3 GAB-CCR分支消融实验结果(%)
方法 准确率 敏感度 特异性 F1-Score Baseline 78.23 61.02 90.25 60.05 Baseline+GAB-SCCR 92.52 87.77 97.83 89.08 Baseline+GAB-DCCR 92.46 86.70 97.73 88.73 Baseline+GAB-CCR 93.48 88.29 98.02 90.78 表 4 消融实验结果(%)
方法 准确率 敏感度 特异性 F1-Score Baseline 78.23 61.02 90.25 60.05 Baseline+GAB-CCR 93.48 88.29 98.02 90.78 Baseline+BFB-WSI 91.91 88.28 97.87 87.17 Baseline+BAB-WSI 93.12 88.34 98.03 89.68 Basline+GAB-CCR+BFB-WSI 93.69 89.50 98.14 91.05 Basline+GAB-CCR+BAB-WSI(Ours) 94.13 91.34 98.36 91.67 -
[1] SINGH D, VIGNAT J, LORENZONI V, et al. Global estimates of incidence and mortality of cervical cancer in 2020: A baseline analysis of the WHO global cervical cancer elimination initiative[J]. The Lancet Global Health, 2023, 11(2): e197–e206. doi: 10.1016/S2214-109X(22)00501-0. [2] KOSS L G. The Papanicolaou test for cervical cancer detection: A triumph and a tragedy[J]. JAMA, 1989, 261(5): 737–743. doi: 10.1001/jama.1989.03420050087046. [3] JUSMAN Y, SARI B P, and RIYADI S. Cervical precancerous classification system based on texture features and support vector machine[C]. 2021 1st International Conference on Electronic and Electrical Engineering and Intelligent System (ICE3IS), Yogyakarta, Indonesia, 2021: 29–33. doi: 10.1109/ICE3IS54102.2021.9649687. [4] MALLI P K and NANDYAL S. Machine learning technique for detection of cervical cancer using k-NN and artificial neural network[J]. International Journal of Emerging Trends & Technology in Computer Science (IJETTCS), 2017, 6(4): 145–149. [5] SUN Guanglu, LI Shaobo, CAO Yanzhen, et al. Cervical cancer diagnosis based on random forest[J]. International Journal of Performability Engineering, 2017, 13(4): 446–457. doi: 10.23940/ijpe.17.04.p12.446457. [6] DONG Na, ZHAI Mengdie, ZHAO Li, et al. Cervical cell classification based on the CART feature selection algorithm[J]. Journal of Ambient Intelligence and Humanized Computing, 2021, 12(2): 1837–1849. doi: 10.1007/s12652-020-02256-9. [7] ARORA A, TRIPATHI A, and BHAN A. Classification of cervical cancer detection using machine learning algorithms[C]. 2021 6th International Conference on Inventive Computation Technologies (ICICT), Coimbatore, India, 2021: 827–835. doi: 10.1109/ICICT50816.2021.9358570. [8] PRUM S, HANDAYANI D O D, and BOURSIER P. Abnormal cervical cell detection using hog descriptor and SVM classifier[C]. 2018 Fourth International Conference on Advances in Computing, Communication & Automation (ICACCA), Subang Jaya, Malaysia, 2018: 1–6. doi: 10.1109/ICACCAF.2018.8776766. [9] QIN Jian, HE Yongjun, GE Jinping, et al. A multi-task feature fusion model for cervical cell classification[J]. IEEE Journal of Biomedical and Health Informatics, 2022, 26(9): 4668–4678. doi: 10.1109/JBHI.2022.3180989. [10] DONG N, ZHAO L, WU C H, et al. Inception v3 based cervical cell classification combined with artificially extracted features[J]. Applied Soft Computing, 2020, 93: 106311. doi: 10.1016/j.asoc.2020.106311. [11] SZEGEDY C, VANHOUCKE V, IOFFE S, et al. Rethinking the inception architecture for computer vision[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 2818–2826. doi: 10.1109/CVPR.2016.308. [12] 赵司琦, 梁義钦, 秦健, 等. 基于样本基准值的宫颈异常细胞识别方法[J]. 哈尔滨理工大学学报, 2022, 27(6): 103–114. doi: 10.15938/j.jhust.2022.06.013.ZHAO Siqi, LIANG Yiqin, QIN Jian, et al. A method for identifying cervical abnormal cells based on sample benchmark values[J]. Journal of Harbin University of Science and Technology, 2022, 27(6): 103–114. doi: 10.15938/j.jhust.2022.06.013. [13] LIN Haoming, HU Yuyang, CHEN Siping, et al. Fine-grained classification of cervical cells using morphological and appearance based convolutional neural networks[J]. IEEE Access, 2019, 7: 71541–71549. doi: 10.1109/ACCESS.2019.2919390. [14] ZHANG Ling, LU Le, NOGUES I, et al. DeepPap: Deep convolutional networks for cervical cell classification[J]. IEEE Journal of Biomedical and Health Informatics, 2017, 21(6): 1633–1643. doi: 10.1109/JBHI.2017.2705583. [15] SHI Jun, WANG Ruoyu, ZHENG Yushan, et al. Cervical cell classification with graph convolutional network[J]. Computer Methods and Programs in Biomedicine, 2021, 198: 105807. doi: 10.1016/j.cmpb.2020.105807. [16] SHANTHI P B, FARUQI F, HAREESHA K S, et al. Deep convolution neural network for malignancy detection and classification in microscopic uterine cervix cell images[J]. Asian Pacific Journal of Cancer Prevention, 2019, 20(11): 3447–3456. doi: 10.31557/APJCP.2019.20.11.3447. [17] MANNA A, KUNDU R, KAPLUN D, et al. A fuzzy rank-based ensemble of CNN models for classification of cervical cytology[J]. Scientific Reports, 2021, 11(1): 14538. doi: 10.1038/s41598-021-93783-8. [18] TRIPATHI A, ARORA A, and BHAN A. Classification of cervical cancer using deep learning algorithm[C]. 2021 5th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 2021: 1210–1218. doi: 10.1109/ICICCS51141.2021.9432382. [19] BASAK H, KUNDU R, CHAKRABORTY S, et al. Cervical cytology classification using PCA and GWO enhanced deep features selection[J]. SN Computer Science, 2021, 2(5): 369. doi: 10.1007/S42979-021-00741-2. [20] LI Jun, DOU Qiyan, YANG Haima, et al. Cervical cell multi-classification algorithm using global context information and attention mechanism[J]. Tissue and Cell, 2022, 74: 101677. doi: 10.1016/j.tice.2021.101677. [21] RAHAMAN M M, LI Chen, YAO Yudong, et al. DeepCervix: A deep learning-based framework for the classification of cervical cells using hybrid deep feature fusion techniques[J]. Computers in Biology and Medicine, 2021, 136: 104649. doi: 10.1016/j.compbiomed.2021.104649. [22] NAYAR R and WILBUR D C, The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes[M]. 3rd ed. Cham: Springer, 2015. doi: 10.1007/978-3-319-11074-5. [23] 郑光远, 刘峡壁, 韩光辉. 医学影像计算机辅助检测与诊断系统综述[J]. 软件学报, 2018, 29(5): 1471–1514. doi: 10.13328/j.cnki.jos.005519.ZHENG Guangyuan, LIU Xiabi, and HAN Guanghui. Survey on medical image computer aided detection and diagnosis systems[J]. Journal of Software, 2018, 29(5): 1471–1514. doi: 10.13328/j.cnki.jos.005519. [24] HOUSSEIN E H, EMAM M M, ALI A A, et al. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review[J]. Expert Systems with Applications, 2021, 167: 114161. doi: 10.1016/j.eswa.2020.114161. [25] GUMMA L N, THIRUVENGATANADHAN R, KURAKULA L, et al. A survey on convolutional neural network (deep-learning technique)-based lung cancer detection[J]. SN Computer Science, 2022, 3(1): 66. doi: 10.1007/S42979-021-00887-Z. [26] 陈皓, 段红柏, 郭紫园, 等. 基于CT征象量化分析的肺结节恶性度分级[J]. 电子与信息学报, 2021, 43(5): 1405–1413. doi: 10.11999/JEIT200167.CHEN Hao, DUAN Hongbai, GUO Ziyuan, et al. Malignancy grading of lung nodules based on CT signs quantization analysis[J]. Journal of Electronics & Information Technology, 2021, 43(5): 1405–1413. doi: 10.11999/JEIT200167. [27] SHAMSHAD F, KHAN S, ZAMIR S W, et al. Transformers in medical imaging: A survey[J]. Medical Image Analysis, 2023, 88: 102802. doi: 10.1016/j.media.2023.102802. [28] HE Kaiming, ZHANG Xiangyu, REN Shaoqing, et al. Deep residual learning for image recognition[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 770–778. doi: 10.1109/CVPR.2016.90. [29] DOSOVITSKIY A, BEYER L, KOLESNIKOV A, et al. An image is worth 16x16 words: Transformers for image recognition at scale[C]. 9th International Conference on Learning Representations, 2021. [30] LIU Wanli, LI Chen, XU Ning, et al. CVM-Cervix: A hybrid cervical Pap-smear image classification framework using CNN, visual transformer and multilayer perceptron[J]. Pattern Recognition, 2022, 130: 108829. doi: 10.1016/j.patcog.2022.108829. [31] DEO B S, PAL M, PANIGRAHI P K, et al. CerviFormer: A pap smear-based cervical cancer classification method using cross-attention and latent transformer[J]. International Journal of Imaging Systems and Technology, 2024, 34(2): e23043. doi: 10.1002/ima.23043. [32] HEMALATHA K, VETRISELVI V, DHANDAPANI M, et al. CervixFuzzyFusion for cervical cancer cell image classification[J]. Biomedical Signal Processing and Control, 2023, 85: 104920. doi: 10.1016/j.bspc.2023.104920. [33] KIPF T N and WELLING M. Semi-supervised classification with graph convolutional networks[C]. 5th International Conference on Learning Representations, Toulon, France, 2017. [34] VELIČKOVIĆ P, CUCURULL G, CASANOVA A, et al. Graph attention networks[J]. arXiv: 1710.10903, 2017. doi: 10.48550/arXiv.1710.10903. [35] LI Yujia, TARLOW D, BROCKSCHMIDT M, et al. Gated graph sequence neural networks[C]. 4th International Conference on Learning Representations, San Juan, Puerto Rico, 2016. [36] ZHOU Jie, CUI Ganqu, HU Shengding, et al. Graph neural networks: A review of methods and applications[J]. AI Open, 2020, 1: 57–81. doi: 10.1016/j.aiopen.2021.01.001. [37] AHMEDT-ARISTIZABAL D, ARMIN M A, DENMAN S, et al. A survey on graph-based deep learning for computational histopathology[J]. Computerized Medical Imaging and Graphics, 2022, 95: 102027. doi: 10.1016/j.compmedimag.2021.102027. [38] BESSADOK A, MAHJOUB M A, and REKIK I. Graph neural networks in network neuroscience[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2023, 45(5): 5833–5848. doi: 10.1109/TPAMI.2022.3209686. [39] ZHANG Xiaomeng, LIANG Li, LIU Lin, et al. Graph neural networks and their current applications in bioinformatics[J]. Frontiers in Genetics, 2021, 12: 690049. doi: 10.3389/fgene.2021.690049. [40] OU Yanglan, XUE Yuan, YUAN Ye, et al. Semi-supervised cervical dysplasia classification with learnable graph convolutional network[C]. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, USA, 2020: 1720–1724. doi: 10.1109/ISBI45749.2020.9098507. [41] DENG Jia, DONG Wei, SOCHER R, et al. ImageNet: A large-scale hierarchical image database[C]. 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, USA, 2009: 248–255. doi: 10.1109/CVPR.2009.5206848. [42] LIU Ze, LIN Yutong, CAO Yue, et al. Swin transformer: Hierarchical vision transformer using shifted windows[C]. Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 9992–10002. doi: 10.1109/ICCV48922.2021.00986. [43] BRODY S, ALON U, and YAHAV E. How attentive are graph attention networks?[C]. The 10th International Conference on Learning Representations, 2022. [44] TAN Mingxing and LE Q V. EfficientNetV2: Smaller models and faster training[C]. Proceedings of the 38th International Conference on Machine Learning, 2021: 10096–10106. [45] WANG Minjie, ZHENG Da, YE Zihao, et al. Deep graph library: A graph-centric, highly-performant package for graph neural networks[J]. arXiv: 1909.01315, 2019. doi: 10.48550/arXiv.1909.01315. [46] WIGHTMAN R. PyTorch-Image-Models[EB/OL]. https://github.com/rwightman/pytorch-image-models, 2021. [47] HUANG Gao, LIU Zhuang, VAN DER MAATEN L, et al. Densely connected convolutional networks[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 2261–2269. doi: 10.1109/CVPR.2017.243. [48] YUAN Li, HOU Qibin, JIANG Zihang, et al. VOLO: Vision outlooker for visual recognition[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2023, 45(5): 6575–6586. doi: 10.1109/TPAMI.2022.3206108. [49] EL-NOUBY A, TOUVRON H, CARON M, et al. XCiT: Cross-covariance image transformers[C]. Proceedings of the 35th International Conference on Neural Information Processing Systems, 2021: 1531. [50] TOLSTIKHIN I, HOULSBY N, KOLESNIKOV A, et al. MLP-mixer: An all-MLP architecture for vision[C]. Proceedings of the 35th International Conference on Neural Information Processing Systems, 2021: 1857. -





 下载:
下载:


 下载:
下载:
