A Dual-path Network Chest Film Disease Classification Method Combined with a Triple Attention Mechanism
-
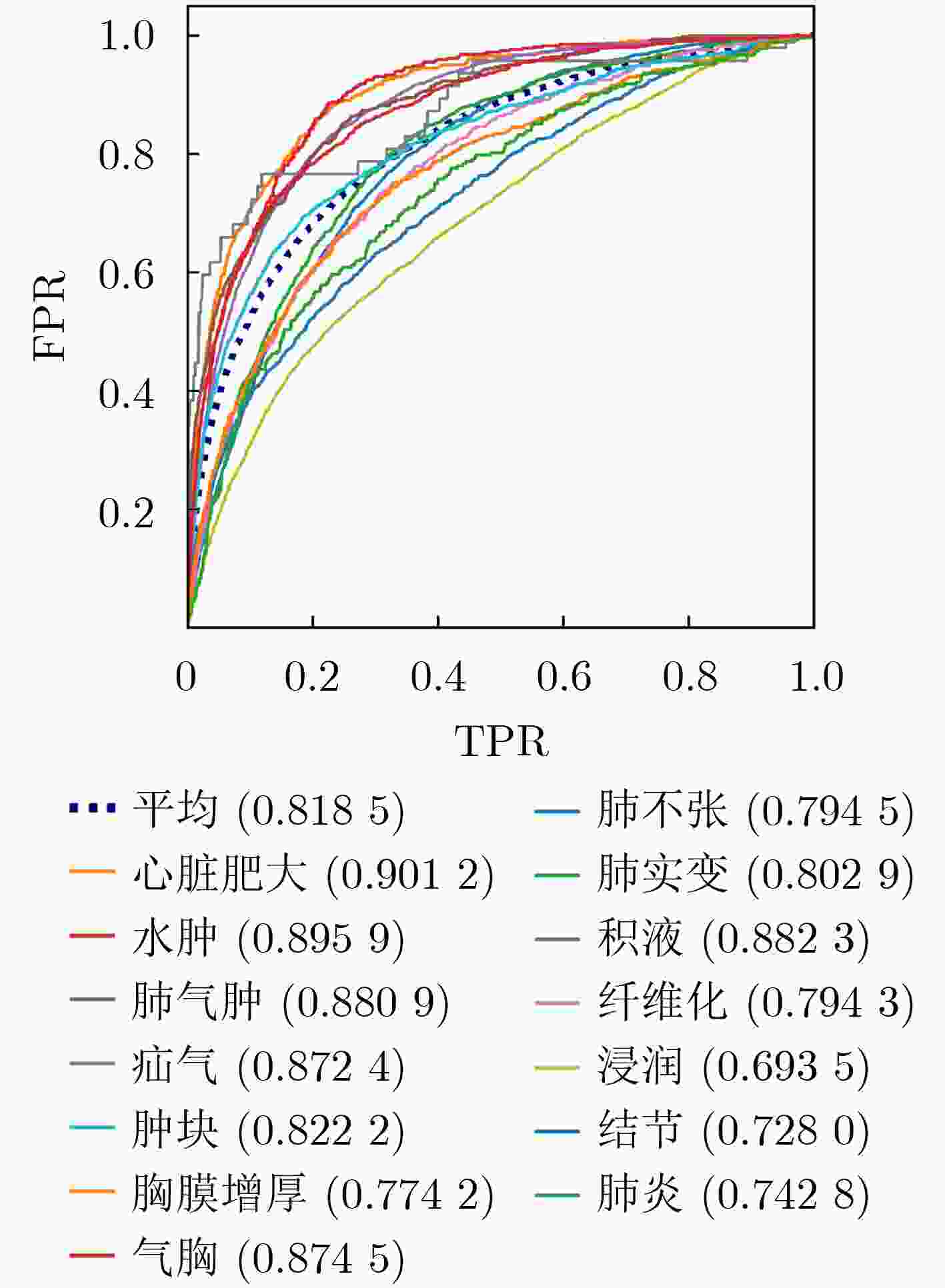
摘要: 近年来,利用CNN进行医学图像处理,在胸片疾病分类任务中取得显著研究进展。然而,与单一结构CNN相比,双路径网络可结合不同CNN特点,从而提高疾病分类能力。其次,对于不同疾病,其位置、大小、形态、密度、纹理等特征均有不同,而注意力机制有助于模型提取不同病理特征,提升分类精度。因此针对胸片疾病分类问题,该文提出一种结合三重注意力机制的双路径卷积神经网络(TADPN),TADPN将ResNet和DenseNet结合的双路径网络DPN作为骨干网络,并利用3种不同形式的注意力机制改进DPN,在维持参数量稳定的同时提高网络复杂度,进而提升对胸片疾病的分类精度。在ChestXray14数据集上实验,并与目前较为先进的6种算法对比,14种疾病的平均AUC值达到0.8185,较前人提升1.1%,表明双路径CNN及三重注意力机制对胸片疾病分类的有效性及TADPN的先进性。Abstract: In recent years, medical image processing with CNN has made remarkable research progress in the task of chest film disease classification. However, compared with single structure CNN, dual-path network can combine the characteristics of different CNN to improve the ability of disease classification. Secondly, for different diseases, their location, size, shape, and texture are different, the attention mechanism helps the model to extract different pathological features and improve the classification accuracy. Therefore, focusing on the chest film disease classification problem, a dual path convolution neural network TADPN(Triple Attention Dual Path Network) combined with a triple attention mechanism is proposed. TADPN takes the dual-path network combined with ResNet and DenseNet as the backbone network and uses three different forms of attention mechanisms to improve the backbone network. The network complexity and classification accuracy are improved while maintaining the stability of the parameters. In this paper, the validity of TADPN is compared with the six advanced algorithms on the ChestXray14 dataset. The experiments show the progressiveness of the dual-path CNN and the triple attention mechanism, as well as the effectiveness of TADPN. The average AUC value of 14 diseases reaches 0.8185, which is 1.1% higher than that of previous generations.
-
Key words:
- Medical image processing /
- Chest film classification /
- CNN /
- Attention mechanism
-
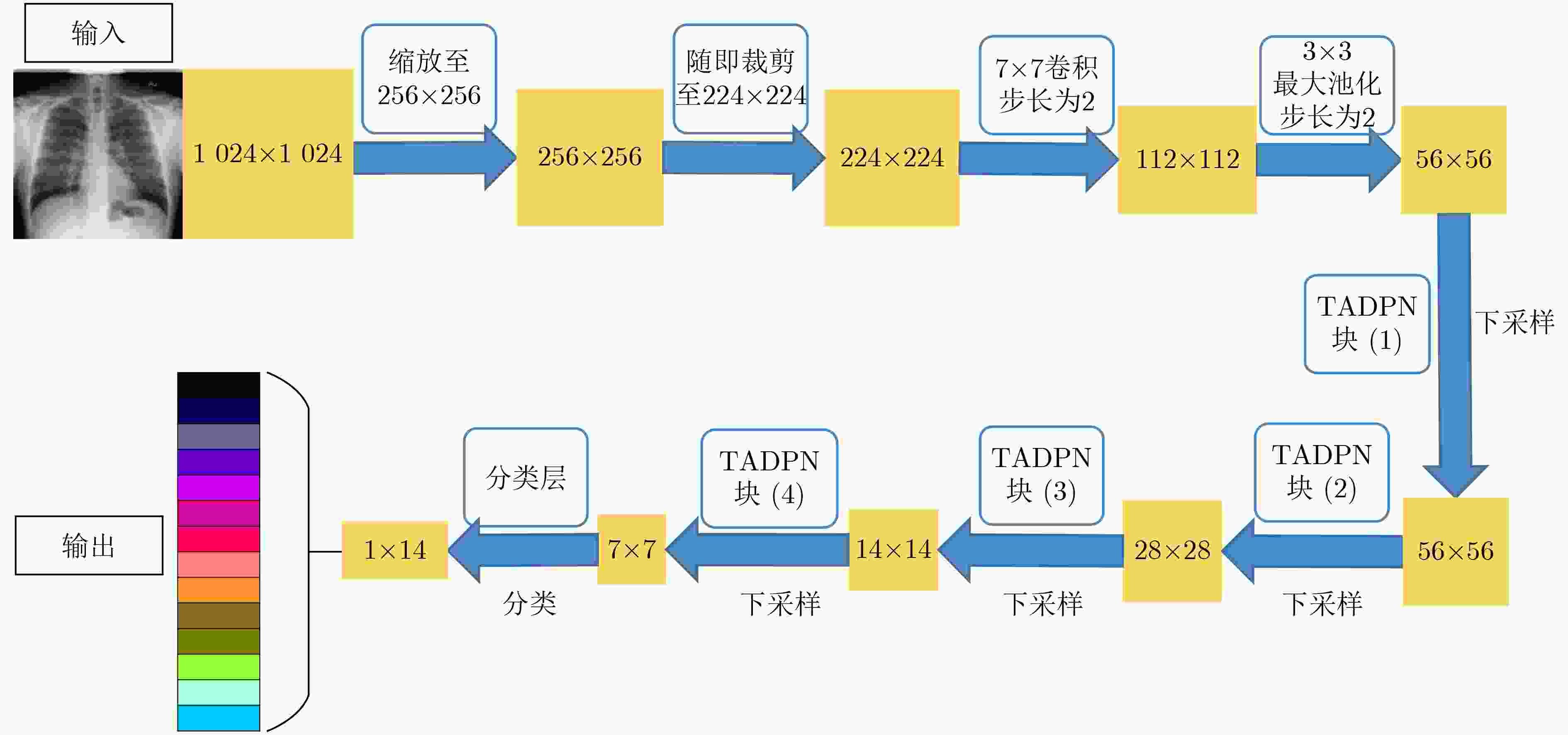
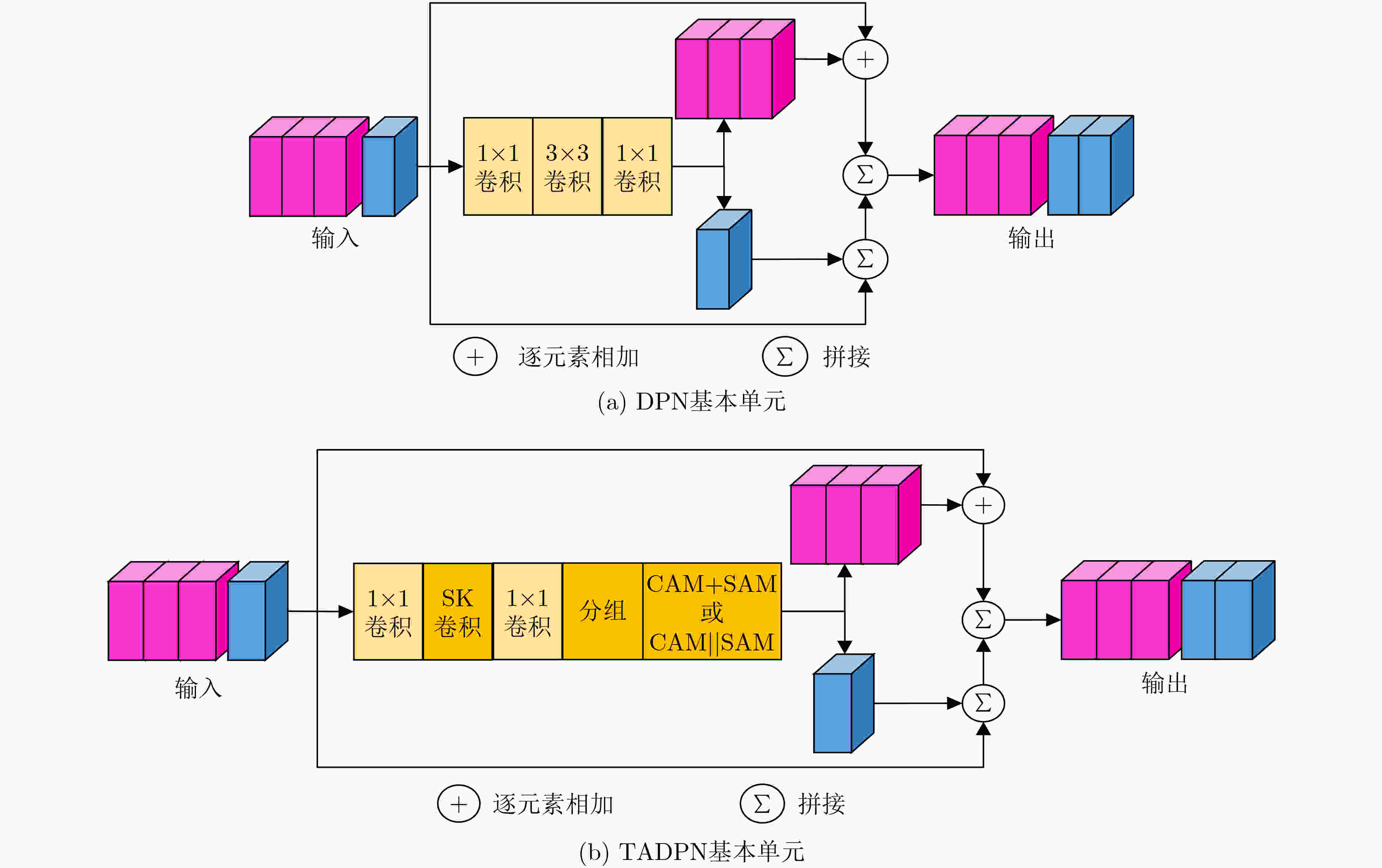
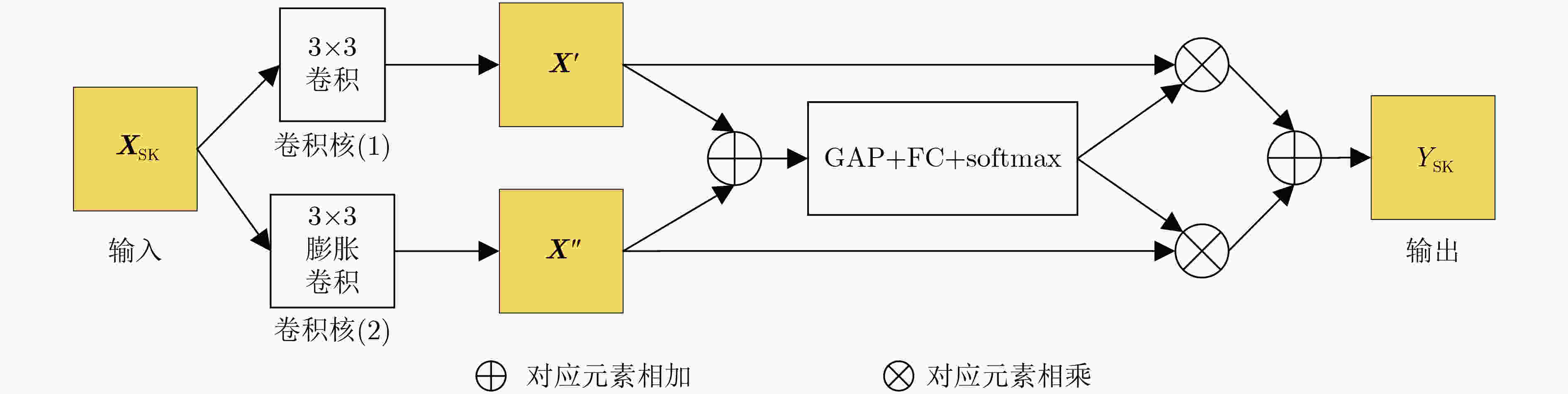
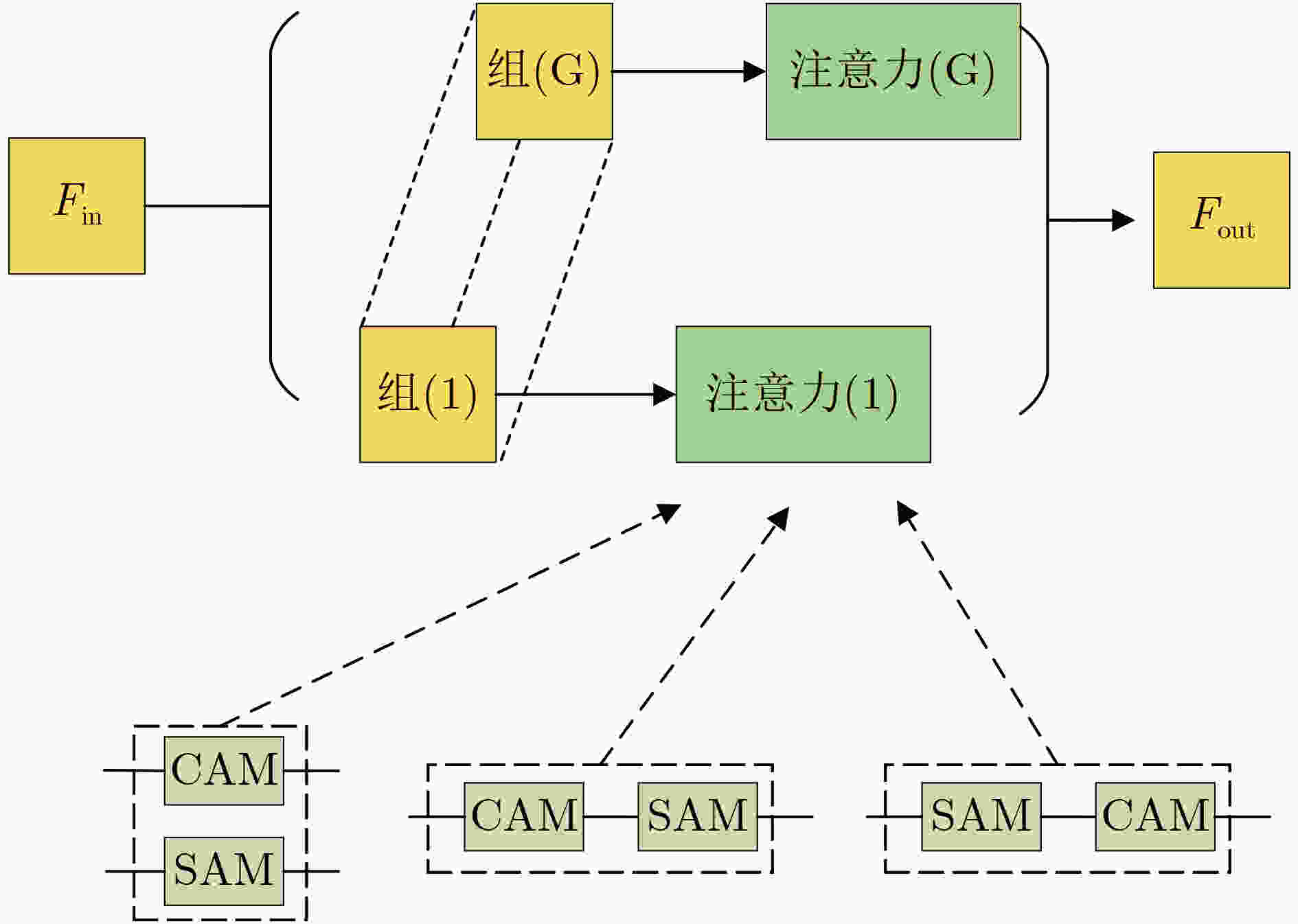
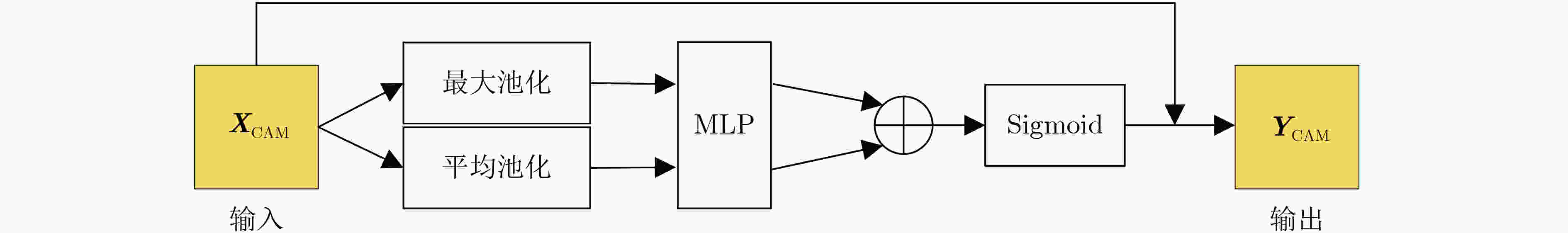
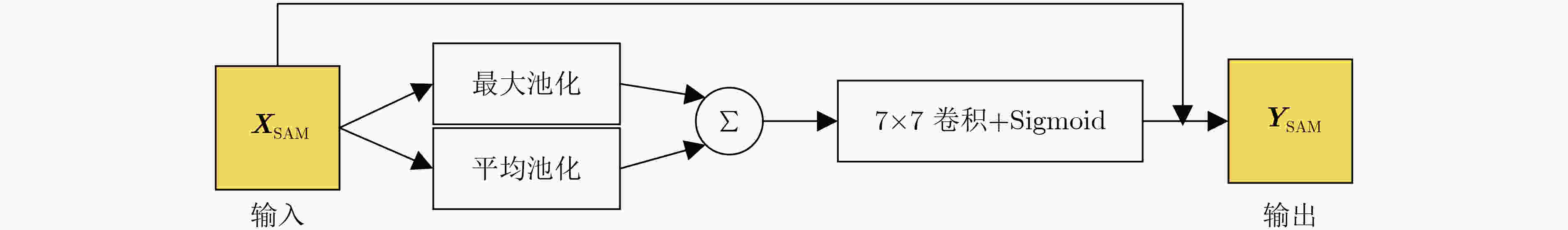
表 1 TADPN网络结构
层 操作 输出尺寸 卷积层 7×7卷积,步长为2 112×112 池化层 3×3最大池化,步长为2 56×56 TADPN块(1) $ \left\{\begin{array}{l}1\times \text{1}卷积、\text{3}\times \text{3 SK}卷积、1\times \text{1}卷积\\ \text{ CAM}、\text{SAM}\end{array}\right\} $×3 56×56 TADPN块(2) $ \left\{\begin{array}{l}1\times \text{1}卷积、\text{3}\times \text{3 SK}卷积、1\times \text{1}卷积\\ \text{ CAM}、\text{SAM}\end{array}\right\} $×4 28×28 TADPN块(3) $ \left\{\begin{array}{l}1\times \text{1}卷积、\text{3}\times \text{3 SK}卷积、1\times \text{1}卷积\\ \text{ CAM}、\text{SAM}\end{array}\right\} $×20 14×14 TADPN块(4) $ \left\{\begin{array}{l}1\times \text{1}卷积、\text{3}\times \text{3 SK}卷积、1\times \text{1}卷积\\ \text{ CAM}、\text{SAM}\end{array}\right\} $×3 7×7 分类层 14维全连接 1×14 表 2 14种胸片疾病的影像学诊断依据
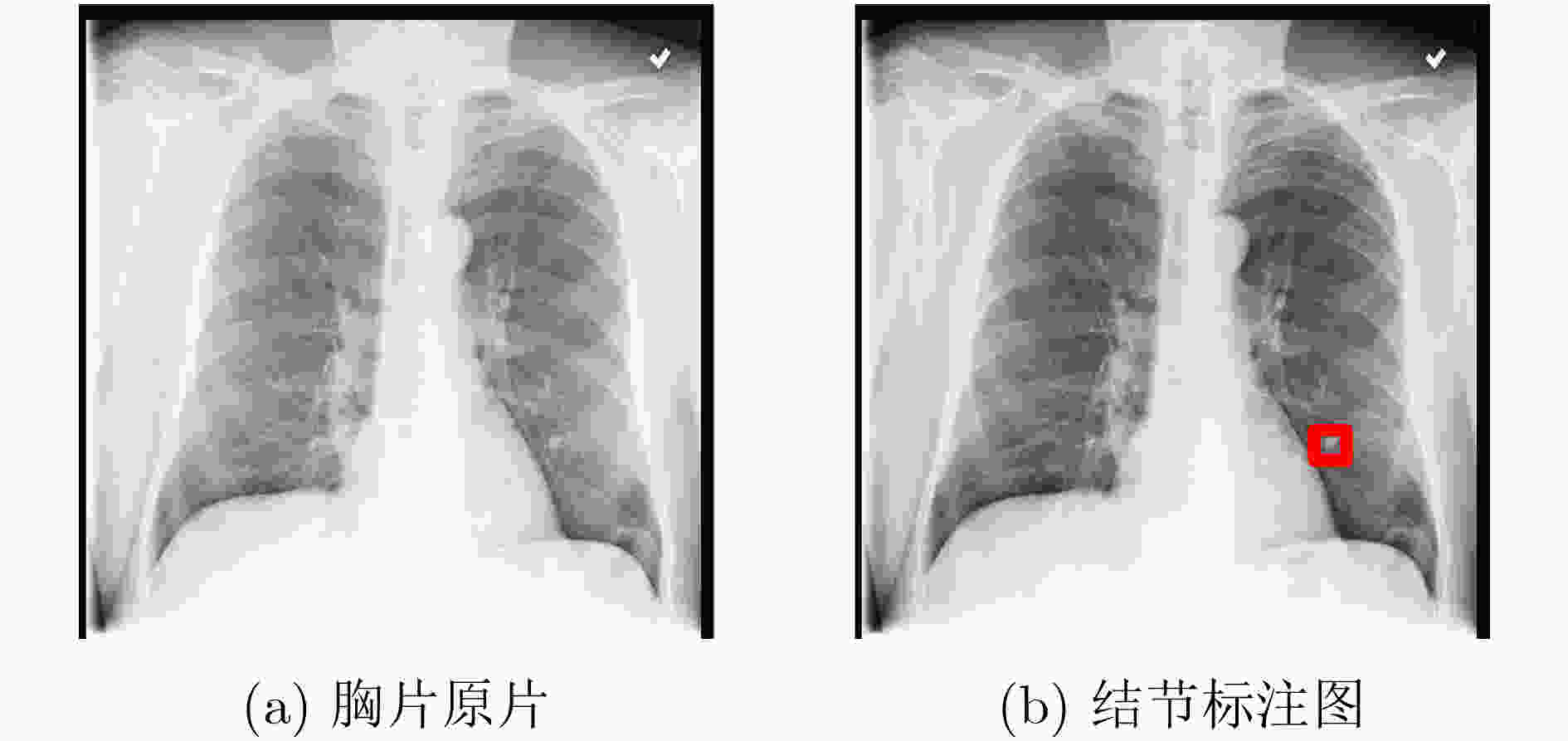
疾病 影像学依据 肺不张 肺野呈均匀致密影,气管、纵隔向患侧移位,肋间隙变窄 心脏肥大 心影增大 积液 肋膈角变钝或消失,纵隔向健侧移位,体液上缘呈外高内低凹面向上弧形影 浸润 患侧浸润性阴影 肿块 肺实质内呈高密度阴影(直径大于3 cm) 结节 肺实质内呈高密度阴影(直径小于3 cm) 肺炎 肺纹理增多、密度增高,呈毛玻璃影,片状模糊,边界不清 气胸 肺被压缩向肺门部收缩,呈均匀无肺纹理走形透亮影 肺实变 呈空气支气管征、肺泡充气征、阴影不透明、血管模糊 水肿 肺纹理以及肺门区血管增粗,肺透亮度降低、肋膈角改变或消失 肺气肿 肺体积增加、纹理增粗、透过度增大 纤维化 肺中下野呈毛玻璃状、典型性改变弥漫性线条状、结节状、云絮样、网状阴影 胸膜增厚 肋隔角变浅、变钝,呈不规则条状钙化 疝气 呈高透光度膨出,与肺组织相连 表 3 ChestXray14数据集疾病种类及数量
疾病名称 数量(张) 疾病名称 数量(张) 疾病名称 数量(张) 肺不张(Atelectasis) 11 559 气胸(Pneumothorax) 5 302 无病 60 361 心脏肥大(Cardiomegaly) 2 776 肺实变(Consolidation) 4 667 积液(Effusion) 13 317 水肿(Edema) 2 303 浸润(Infiltration) 19 894 肺气肿(Emphysema) 2 516 肿块(Mass) 5 782 纤维化(Fibrosis) 1 686 结节(Nodule) 6 331 胸膜增厚(Pleural Thickening) 3 385 肺炎(Pneumonia) 1 431 疝气(Hernia) 227 患病合计 51 759 总计 112 120 表 4 混淆矩阵
预测类别 真实类别 1(患病) 0(无病) 1(患病) 真阳 (TP) 伪阳(FP) 0(无病) 伪阴 (FN) 真阴(TN) 表 5 不同模型在ChestX-ray14数据集上的结果比较
基于ResNet模型 基于DenseNet模型 TADPN 文献[14] 文献[35] 文献[21] 文献[36] 文献[37] 文献[28] 本文 肺不张 0.755 7 0.80 0.800 4 0.762 7 0.795 0.785 0 0.794 5 心脏肥大 0.886 5 0.87 0.879 8 0.883 5 0.887 0.876 6 0.901 2 积液 0.819 1 0.87 0.872 0.815 9 0.875 0.862 8 0.882 3 浸润 0.689 2 0.70 0.712 2 0.678 6 0.703 0.673 0 0.693 5 肿块 0.813 6 0.83 0.795 3 0.801 2 0.83 5 0.804 0 0.822 2 结节 0.754 5 0.75 0.720 5 0.729 3 0.716 0.729 9 0.728 0 肺炎 0.729 2 0.67 0.734 7 0.709 7 0.742 0.742 3 0.742 8 气胸 0.849 9 0.87 0.842 2 0.837 7 0.863 0.842 6 0.874 5 肺实变 0.728 3 0.80 0.801 0.744 3 0.786 0.784 6 0.802 9 水肿 0.847 5 0.88 0.878 7 0.841 4 0.892 0.872 7 0.895 9 肺气肿 0.907 5 0.91 0.853 6 0.883 6 0.875 0.858 0 0.880 9 纤维化 0.817 9 0.78 0.798 0.807 7 0.756 0.775 4 0.794 3 胸膜增厚 0.764 7 0.76 0.743 1 0.753 6 0.774 0.775 6 0.774 2 疝气 0.874 7 0.77 0.871 1 0.876 3 0.836 0.864 5 0.872 4 平均 0.802 7 0.804 0.807 3 0.794 1 0.809 7 0.802 0 0.818 5 表 6 3种骨干网络分类结果对比
肺不张 心脏肥大 积液 浸润 肿块 结节 肺炎 ResNet-101 0.790 0 0.907 7 0.870 5 0.688 2 0.8074 0.720 8 0.706 9 DenseNet-121 0.787 2 0.903 0 0.872 4 0.685 3 0.802 5 0.709 6 0.722 2 DPN-92 0.791 3 0.910 6 0.873 4 0.684 2 0.807 6 0.713 8 0.730 1 气胸 肺实变 水肿 肺气肿 纤维化 胸膜增厚 疝气 平均 ResNet-101 0.860 4 0.794 8 0.890 0 0.868 2 0.777 4 0.761 1 0.864 5 0.807 7 DenseNet-121 0.861 6 0.799 4 0.886 3 0.866 0 0.773 9 0.763 2 0.895 1 0.809 1 DPN-92 0.861 5 0.797 9 0.888 8 0.867 7 0.782 7 0.774 6 0.897 4 0.813 0 表 7 消融结果对比
肺不张 心脏肥大 积液 浸润 肿块 结节 肺炎 骨干模型 0.791 3 0.910 6 0.873 4 0.684 2 0.807 6 0.713 8 0.730 1 移除SK 0.787 4 0.910 2 0.878 9 0.690 8 0.808 7 0.720 4 0.743 5 移除CAM 0.791 9 0.907 4 0.875 0 0.684 6 0.815 3 0.722 3 0.736 9 移除SAM 0.786 4 0.907 4 0.875 8 0.688 7 0.816 4 0.721 7 0.738 6 TADPN 0.794 5 0.901 2 0.882 3 0.693 5 0.822 2 0.728 0 0.742 8 气胸 肺实变 水肿 肺气肿 纤维化 胸膜增厚 疝气 平均 骨干模型 0.861 5 0.797 9 0.888 8 0.867 7 0.782 7 0.774 6 0.897 4 0.813 0 移除SK 0.865 9 0.804 6 0.897 1 0.879 3 0.783 4 0.770 2 0.875 0 0.815 4 移除CAM 0.867 2 0.802 7 0.894 2 0.881 2 0.775 8 0.774 4 0.890 0 0.815 6 移除SAM 0.873 2 0.803 4 0.887 5 0.876 1 0.783 6 0.780 5 0.872 0 0.815 1 TADPN 0.874 5 0.802 9 0.895 9 0.880 9 0.794 3 0.774 2 0.872 4 0.818 5 表 8 SK卷积参数设置对比
分支1 分支2 平均AUC Parameters(M) FLOPs(G) 卷积核1 膨胀系数D1 卷积核2 膨胀系数D2 3×3 1 3×3 2 0.818 5 4.565 1 20.579 4 3×3 1 3×3 3 0.817 9 4.565 1 20.579 4 3×3 2 3×3 3 0.817 7 4.565 1 20.579 4 3×3 1 5×5 1 0.817 1 4.973 0 22.534 7 3×3 1 7×7 1 0.815 5 5.584 8 25.467 7 5×5 1 5×5 2 0.814 0 5.380 8 24.490 1 5×5 1 7×7 1 0.812 2 5.992 7 27.423 1 表 9 注意力模块组合结果对比
模型 平均AUC Parameters(M) FLOPs(G) DPN-92 0.813 0 17.109 5 4.315 1 DPN-92+SK 0.814 4 19.257 1 4.560 8 DPN-92+CAM||SAM 0.815 0 18.431 8 4.319 3 DPN-92+CAM+SAM 0.815 4 18.431 8 4.319 3 DPN-92+SAM+CAM 0.814 8 18.431 8 4.319 3 TADPN 0.818 5 20.579 4 4.565 1 -
[1] BALA D. Childhood pneumonia recognition using convolutional neural network from chest X-ray images[J]. Journal of Electrical Engineering, Electronics, Control and Computer Science, 2021, 7(26): 33–40. [2] World Health Organization. Causes of death among children[EB/OL]. https://www.who.int/maternal_child_adolescent/data/causes-death-children/en/.2018.1. [3] KHOBRAGADE S, TIWARI A, PATIL C Y, et al. Automatic detection of major lung diseases using Chest Radiographs and classification by feed-forward artificial neural network[C]. 2016 IEEE 1st International Conference on Power Electronics, Intelligent Control and Energy Systems, Delhi, India, 2016: 1–5. [4] QIN Chunli, YAO Demin, SHI Yonghong, et al. Computer-aided detection in chest radiography based on artificial intelligence: A survey[J]. Biomedical Engineering Online, 2018, 17(1): 113. doi: 10.1186/s12938-018-0544-y [5] AYESHA H, IQBAl S, TARIQ M, et al. Automatic medical image interpretation: State of the art and future directions[J]. Pattern Recognition, 2021, 114: 107856. doi: 10.1016/j.patcog.2021.107856 [6] PEZZANO G, RIPOLL V R, and RADEVA P. CoLe-CNN: Context-learning convolutional neural network with adaptive loss function for lung nodule segmentation[J]. Computer Methods and Programs in Biomedicine, 2021, 198: 105792. doi: 10.1016/j.cmpb.2020.105792 [7] 赵奕名, 李锵, 关欣. 组卷积轻量级脑肿瘤分割网络[J]. 中国图象图形学报, 2020, 25(10): 2159–2170. doi: 10.11834/jig.200247ZHAO Yiming, LI Qiang, and GUAN Xin. Lightweight brain tumor segmentation algorithm based on a group convolutional neural network[J]. Journal of Image and Graphics, 2020, 25(10): 2159–2170. doi: 10.11834/jig.200247 [8] 明涛, 王丹, 郭继昌, 等. 基于多尺度通道重校准的乳腺癌病理图像分类[J]. 浙江大学学报:工学版, 2020, 54(7): 1289–1297. doi: 10.3785/j.issn.1008-973X.2020.07.006MING Tao, WANG Dan, GUO Jichang, et al. Breast cancer histopathological image classification using multi-scale channel squeeze-and-excitation model[J]. Journal of Zhejiang University:Engineering Science, 2020, 54(7): 1289–1297. doi: 10.3785/j.issn.1008-973X.2020.07.006 [9] TEIXEIRA V, BRAZ L, PEDRINI H, et al. DuaLAnet: Dual lesion attention network for thoracic disease classification in chest X-rays[C]. 2020 International Conference on Systems, Signals and Image Processing, Niteroi, Brazil, 2020: 69–74. [10] LUO Luyang, YU Lequan, CHEN Hao, et al. Deep mining external imperfect data for chest X-ray disease screening[J]. IEEE Transactions on Medical Imaging, 2020, 39(11): 3583–3594. doi: 10.1109/TMI.2020.3000949 [11] PANT H, LOHANI M C, BHATT A K, et al. Thoracic disease detection using deep learning[C]. 2021 5th International Conference on Computing Methodologies and Communication, Erode, India, 2021: 1197–1203. [12] HE Kaiming, ZHANG Xiangyu, REN Shaoqing, et al. Deep residual learning for image recognition[C]. The 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 2016: 770–778. [13] OUYANG Xi, KARANAM S, WU Ziyan, et al. Learning hierarchical attention for weakly-supervised chest X-ray abnormality localization and diagnosis[J]. IEEE Transactions on Medical Imaging, 2021, 40(10): 2698–2710. doi: 10.1109/TMI.2020.3042773 [14] TANG Yuxing, WANG Xiaosong, HARRISON A P, et al. Attention-guided curriculum learning for weakly supervised classification and localization of thoracic diseases on chest radiographs[C]. The 9th International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 2018: 249–258. [15] HUANG Gao, LIU Zhuang, VAN DER MAATEN L, et al. Densely connected convolutional networks[C]. 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 2261–2269. [16] WANG Hao, YANG Yuanyuan, PAN Yang, et al. Detecting thoracic diseases via representation learning with adaptive sampling[J]. Neurocomputing, 2020, 406: 354–360. doi: 10.1016/j.neucom.2019.06.113 [17] GÜNDEL S, SETIO A A A, GHESU F C, et al. Robust classification from noisy labels: Integrating additional knowledge for chest radiography abnormality assessment[J]. Medical Image Analysis, 2021, 72: 102087. doi: 10.1016/j.media.2021.102087 [18] GUAN Qingji, HUANG Yaping, LUO Yawei, et al. Discriminative feature learning for thorax disease classification in chest X-ray images[J]. IEEE Transactions on Image Processing, 2021, 30: 2476–2487. doi: 10.1109/TIP.2021.3052711 [19] 黄欣, 方钰, 顾梦丹. 基于卷积神经网络的X线胸片疾病分类研究[J]. 系统仿真学报, 2020, 32(6): 1188–1194. doi: 10.16182/j.issn1004731x.joss.18-0712HUANG Xin, FANG Yu, and GU Mengdan. Classification of chest X-ray disease based on convolutional neural network[J]. Journal of System Simulation, 2020, 32(6): 1188–1194. doi: 10.16182/j.issn1004731x.joss.18-0712 [20] GÜNDEL S, GRBIC S, GEORGESCU B, et al. Learning to recognize abnormalities in chest x-rays with location-aware dense networks[C]. The 23rd Iberoamerican Congress on Pattern Recognition, Madrid, Spain, 2018: 757–765. [21] YANG Xiaoyilei, XU Shuaijing, WANG Jian, et al. Attention mechanism in radiologist-level thorax diseases detection[J]. Procedia Computer Science, 2020, 174: 524–529. doi: 10.1016/j.procs.2020.06.120 [22] HU Jie, SHEN Li, and SUN Gang. Squeeze-and-excitation networks[C]. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 7132–7141. [23] GUAN Qingji and HUANG Yaping. Multi-label chest X-ray image classification via category-wise residual attention learning[J]. Pattern Recognition Letters, 2020, 130: 259–266. doi: 10.1016/j.patrec.2018.10.027 [24] CHEN Yunpeng, LI Jianan, XIAO Huaxin, et al. Dual path networks[C]. The 31st International Conference on Neural Information Processing Systems, Long Beach, USA, 2017: 4470–4478. [25] LI Xiang, WANG Wenhai, HU Xiaolin, et al. Selective kernel networks[C]. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, USA, 2019: 510–519. [26] WOO S, PARK J, LEE J Y, et al. CBAM: Convolutional block attention module[C]. The 15th European Conference on Computer Vision, Munich, Germany, 2018: 3–19. [27] WANG Xiaosong, PENG Yifan, LU Le, et al. ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases[C]. 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 3462–3471. [28] 张智睿, 李锵, 关欣. 密集挤压激励网络的多标签胸部X光片疾病分类[J]. 中国图象图形学报, 2020, 25(10): 2238–2248. doi: 10.11834/jig.200232ZHANG Zhirui, LI Qiang, and GUAN Xin. Multilabel chest X-ray disease classification based on a dense squeeze-and-excitation network[J]. Journal of Image and Graphics, 2020, 25(10): 2238–2248. doi: 10.11834/jig.200232 [29] 王粉花, 赵波, 黄超, 等. 基于多尺度和注意力融合学习的行人重识别[J]. 电子与信息学报, 2020, 42(12): 3045–3052. doi: 10.11999/JEIT190998WANG Fenhua, ZHAO Bo, HUANG Chao, et al. Person re-identification based on multi-scale network attention fusion[J]. Journal of Electronics &Information Technology, 2020, 42(12): 3045–3052. doi: 10.11999/JEIT190998 [30] 尹梦晓, 林振峰, 杨锋. 基于动态感受野的自适应多尺度信息融合的图像转换[J]. 电子与信息学报, 2021, 43(8): 2386–2394. doi: 10.11999/JEIT200675YIN Mengxiao, LIN Zhenfeng, and YANG Feng. Adaptive multi-scale information fusion based on dynamic receptive field for image-to-image translation[J]. Journal of Electronics &Information Technology, 2021, 43(8): 2386–2394. doi: 10.11999/JEIT200675 [31] 王睿川, 王岩飞. 基于半监督空间-通道选择性卷积核网络的极化SAR图像地物分类[J]. 雷达学报, 2021, 10(4): 516–530. doi: 10.12000/JR21080WANG Ruichuan and WANG Yanfei. Terrain classification of polarimetric SAR images using semi-supervised spatial-channel selective kernel network[J]. Journal of Radars, 2021, 10(4): 516–530. doi: 10.12000/JR21080 [32] XIE Saining, GIRSHICK R, DOLLÁR P, et al. Aggregated residual transformations for deep neural networks[C]. 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 5987–5995. [33] 黎英, 宋佩华. 迁移学习在医学图像分类中的研究进展[J]. 中国图象图形学报, 2022, 27(3): 672–686. doi: 10.11834/jig.210814LI Ying and SONG Peihua. Review of transfer learning in medical image classification[J]. Journal of Image and Graphics, 2022, 27(3): 672–686. doi: 10.11834/jig.210814 [34] 赵晓晴, 李慧盈, 苏安炀, 等. 基于加权损失函数的粘连白细胞分割算法[J]. 吉林大学学报:理学版, 2021, 59(1): 85–91. doi: 10.13413/j.cnki.jdxblxb.2020003ZHAO Xiaoqing, LI Huiying, SU Anyang, et al. Adhesive leukocyte segmentation algorithm based on weighted loss function[J]. Journal of Jilin University:Science Edition, 2021, 59(1): 85–91. doi: 10.13413/j.cnki.jdxblxb.2020003 [35] LI Zhe, WANG Chong, HAN Mei, et al. Thoracic disease identification and localization with limited supervision[C]. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 8290–8299. [36] MA Yanbo, ZHOU Qiuhao, CHEN Xuesong, et al. Multi-attention network for thoracic disease classification and localization[C]. ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing, Brighton, UK, 2019: 1378–1382. [37] HO T K K and GWAK J. Multiple feature integration for classification of thoracic disease in chest radiography[J]. Applied Sciences, 2019, 9(19): 4130. doi: 10.3390/app9194130 [38] PRASHANT P. Chest X-ray (Covid-19 & Pneumonia)[EB/OL]. https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia. [39] CHOWDHURY M E H, RAHMAN T, KHANDAKAR A, et al. Can AI help in screening viral and COVID-19 pneumonia?[J]. IEEE Access, 2020, 8: 132665–132676. doi: 10.1109/ACCESS.2020.3010287 [40] SELVARAJU R R, COGSWELL M, DAS A, et al. Grad-CAM: Visual explanations from deep networks via gradient-based localization[C]. 2017 IEEE International Conference on Computer Vision, Venice, Italy, 2017: 618–626. -





 下载:
下载:

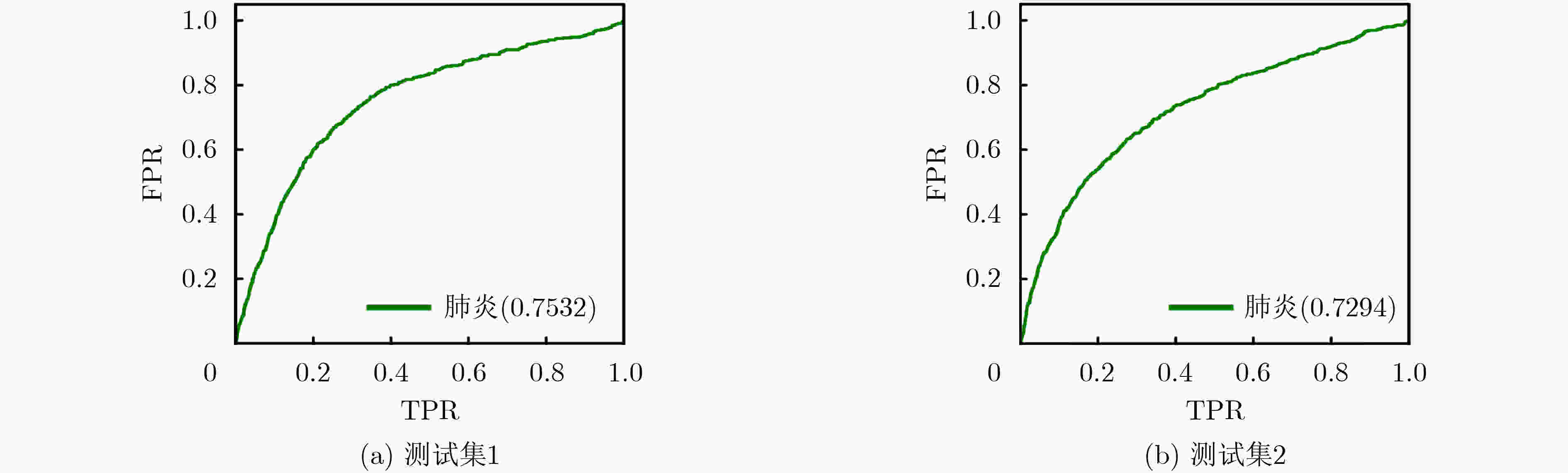
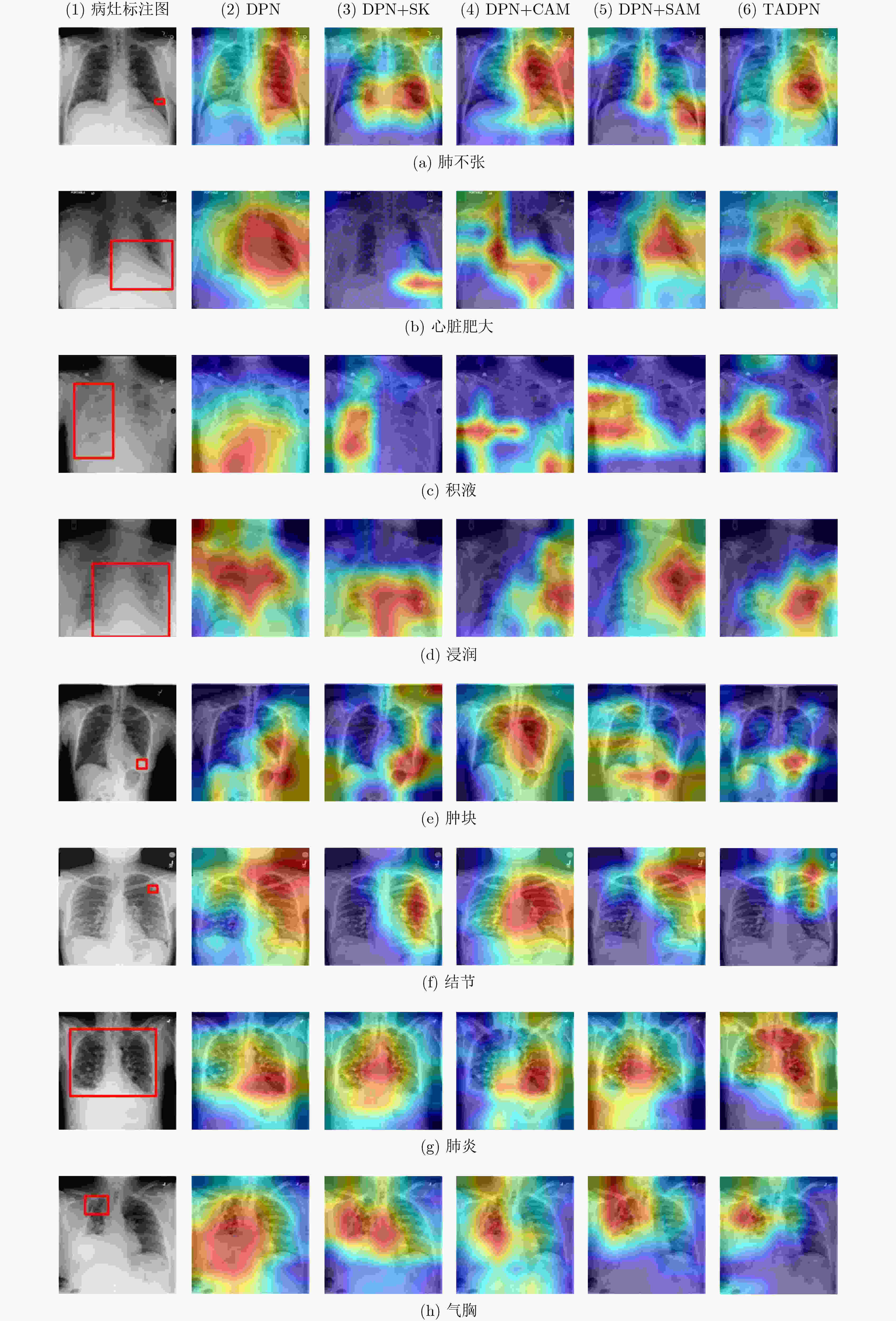


 下载:
下载:
