Polyp Segmentation Using Stair-structured U-Net
-
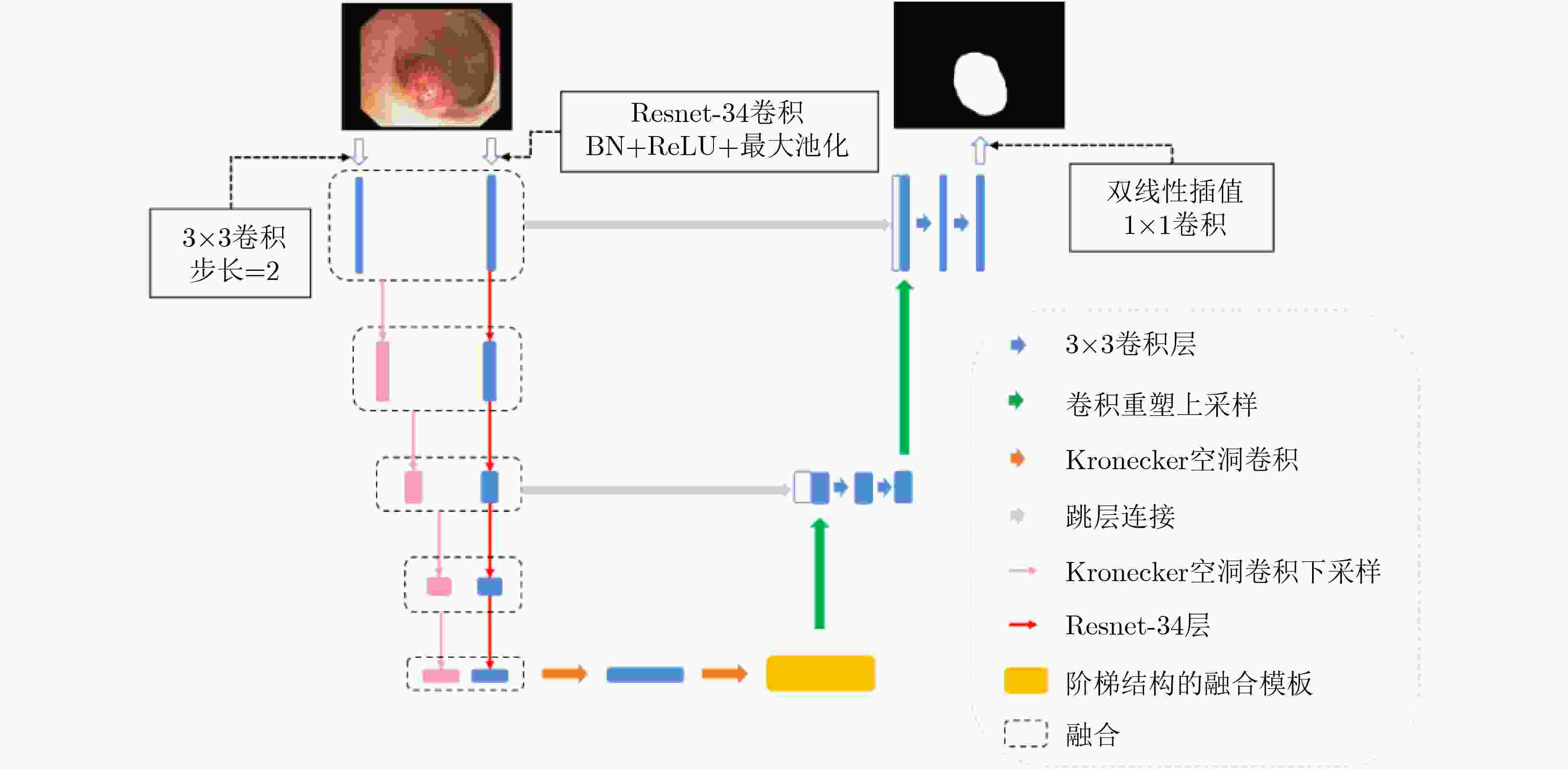
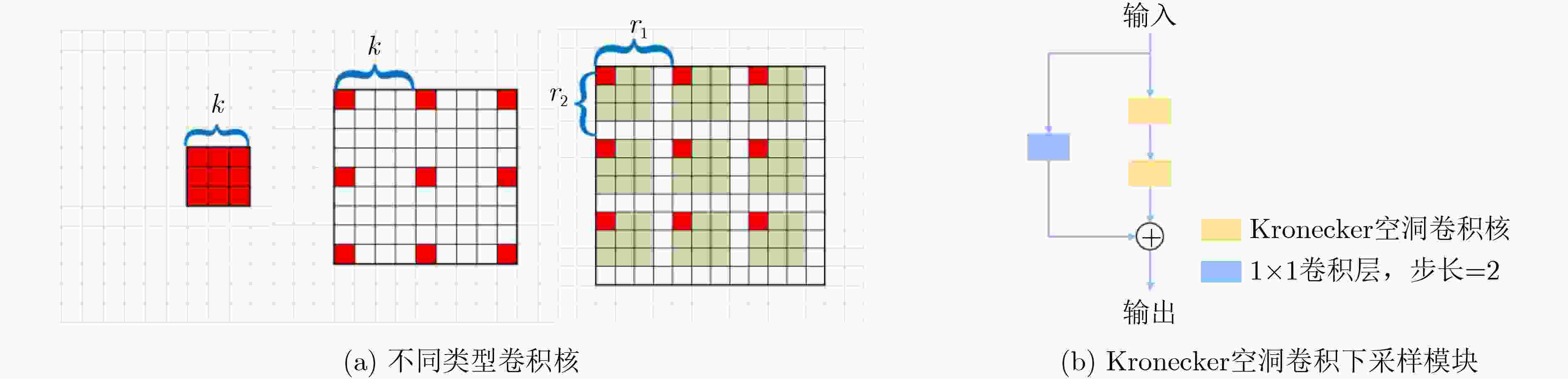
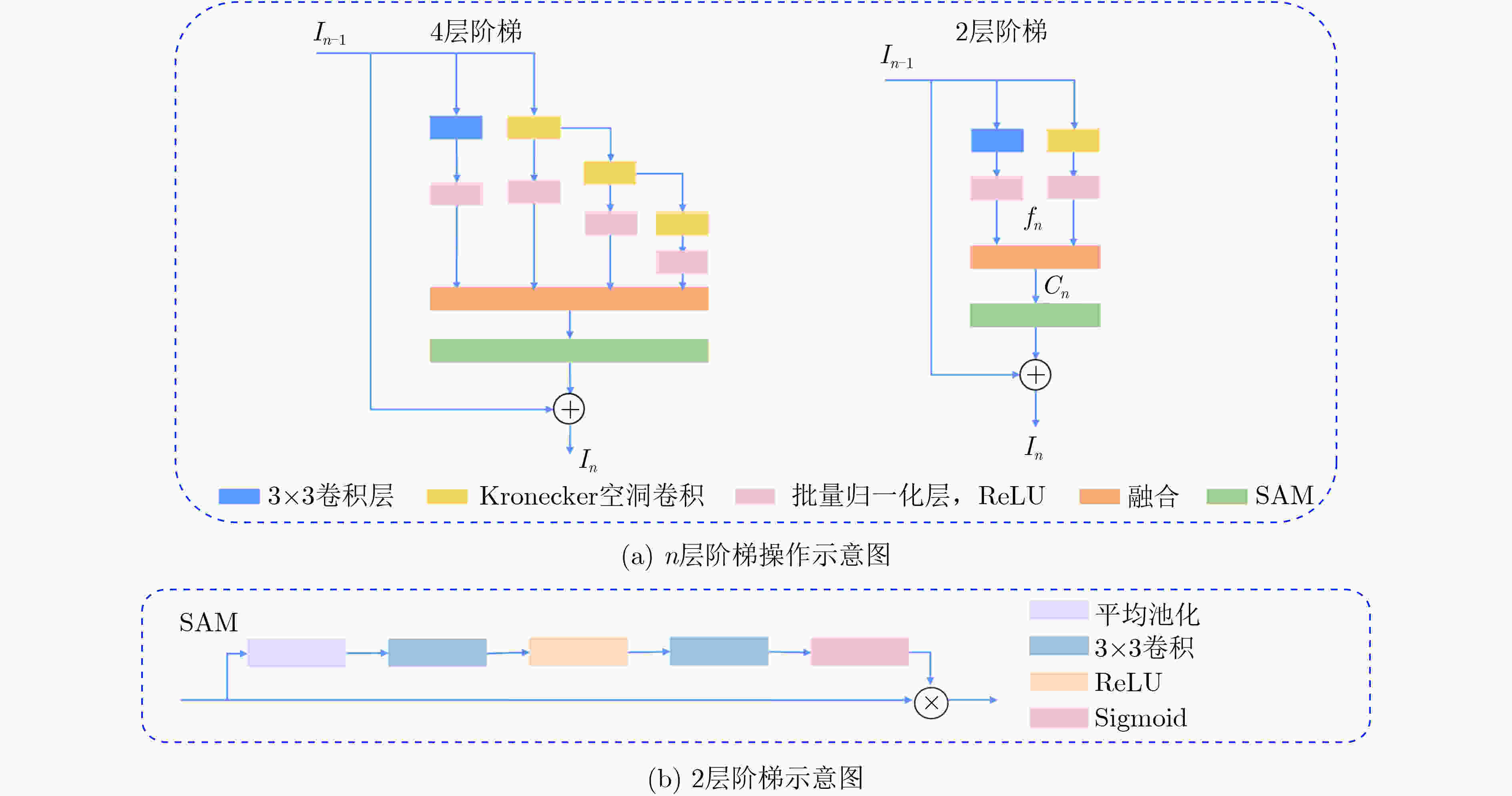
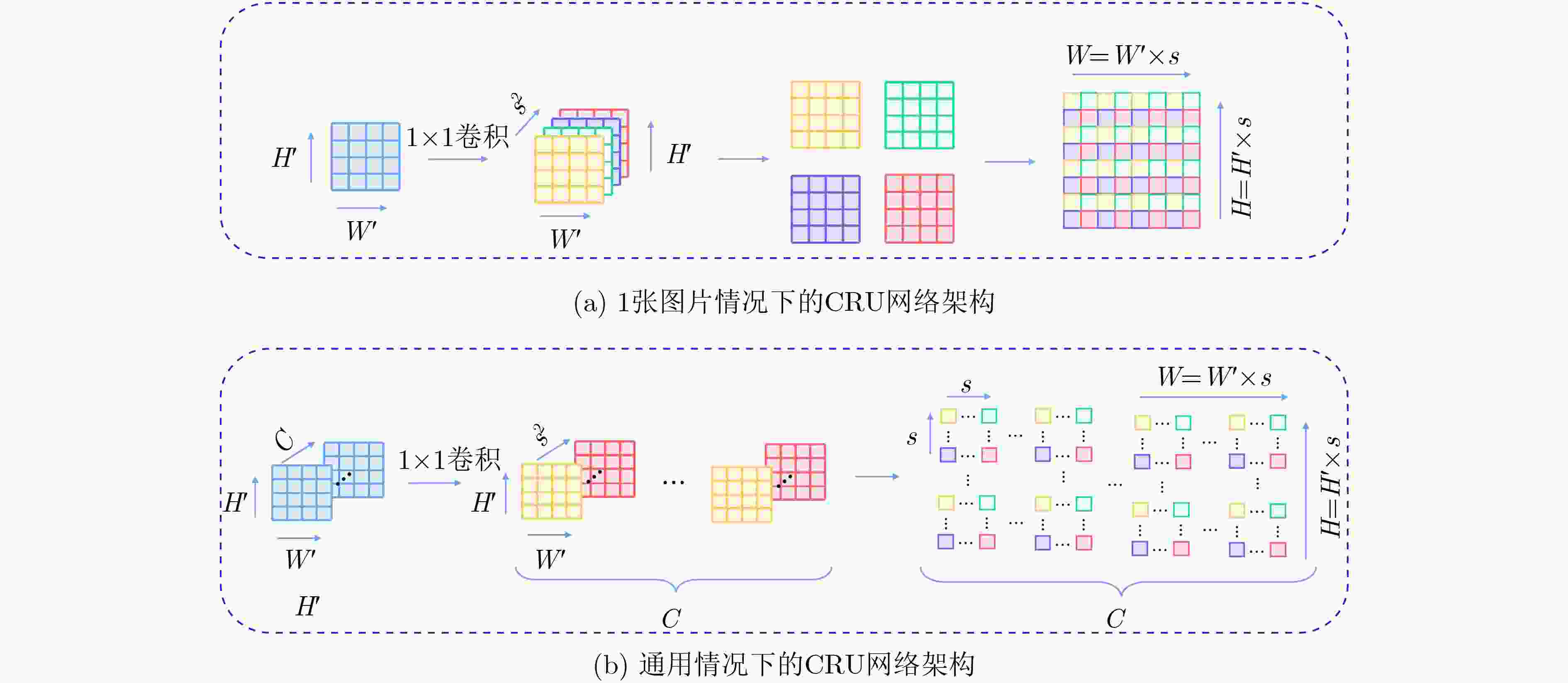
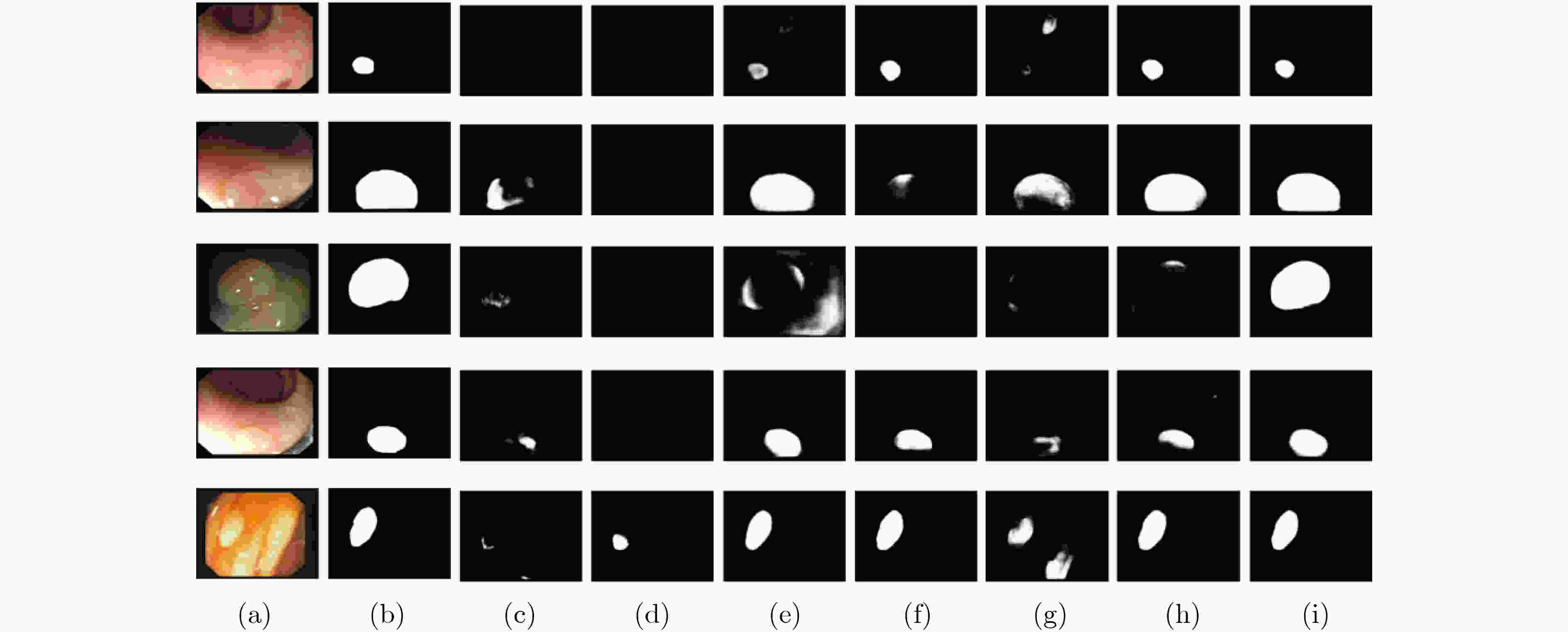
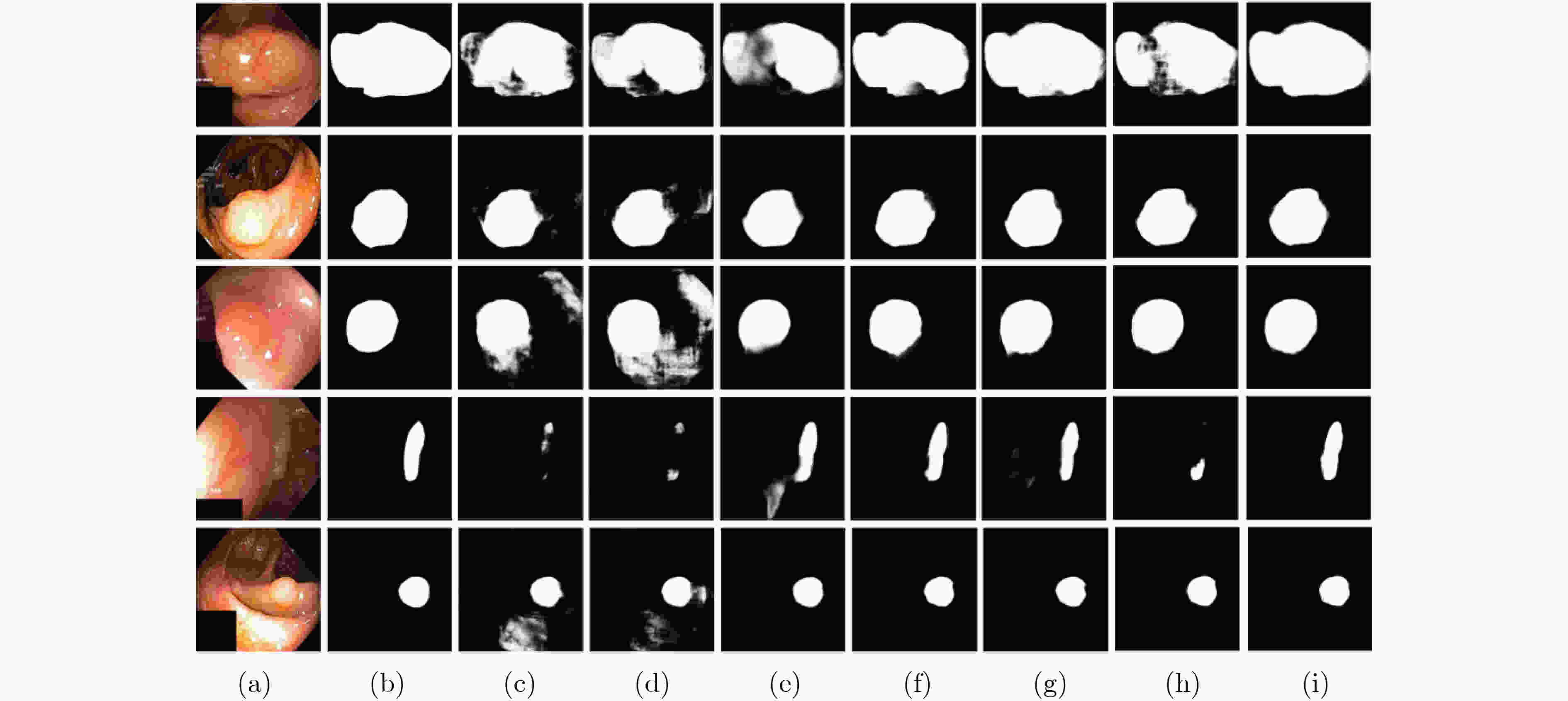
摘要: 结肠息肉的精确分割对结直肠癌的诊断和治疗具有重要意义,目前的分割方法普遍存在有伪影、分割精度低等问题。该文提出一种基于阶梯结构的U-Net结肠息肉分割算法(SU-Net),使用U-Net的U型结构,利用Kronecker乘积来扩展标准空洞卷积核,构成Kronecker空洞卷积下采样有效扩大感受野,弥补传统空洞卷积容易丢失的细节特征;应用具有阶梯结构的融合模块,遵循扩展和堆叠原则形成阶梯状的分层结构,有效捕获上下文信息并从多个尺度聚合特征;在解码器引入卷积重构上采样模块生成密集的像素级预测图,捕获双线性插值上采样中缺少的精细信息。在Kvasir-SEG数据集和CVC-EndoSceneStill数据集上对模型进行了测试,相似系数(Dice)指标和交并比(IoU)指标分别达到了87.51%, 88.75%和82.30%, 85.64%。实验结果表明,该文所提方法改善了因过度曝光、低对比度引起的分割精度低的问题,同时消除了边界外部的图像伪影和图像内部不连贯的现象,优于其他息肉分割方法。Abstract: The precise segmentation of colon polyps plays a significant role in the diagnosis and treatment of colorectal cancer. The existing segmentation methods have generally artifacts and low segmentation accuracy. In this paper, Stair-structured U-Net (SU-Net) is proposed to segment polyp, using U-shaped structure. The Kronecker product is used to extend the standard atrous convolution kernel to keep more detail structrural features that are easily ignored. Stair-structured fusion module is applied to encompass effectively multi-scale features. The decoder introduces a convolutional reshaped upsampling module to generate pixel-level predictions. Experiments are performed on the Kvasir-SEG dataset and the CVC-EndoSceneStill dataset. The results show that the method proposed in this paper outperforms other polyp segmentation methods in Dice and Intersection-over-Union(IoU).
-
Key words:
- Image segmentation /
- Colorectal polyp image /
- Atrous convolution /
- U-Net
-
表 1 SU-Net消融实验列表
序号 实验描述 1 基线 2 仅将基线里的空洞卷积替换为Kronecker空洞卷积 3 将实验2中下采样替换为Kronecker空洞卷积下采样 4 在实验3中的编码器解码器之间加入阶梯结构的融合模块 5 SU-Net 表 2 在EndoSceneStill数据集上各实验的量化结果
评估标准 消融实验编号 1 2 3 4 5 召回率 0.7819 0.8195 0.8028 0.8027 0.8237 特异性 0.9931 0.9908 0.9946 0.9947 0.9929 精确率 0.9185 0.8747 0.9179 0.9119 0.9007 F1 0.7899 0.7994 0.8088 0.8174 0.8230 F2 0.7791 0.8025 0.7980 0.8046 0.8175 IoU 0.7194 0.7214 0.7360 0.7450 0.7499 IoUB 0.9601 0.9599 0.9269 0.9627 0.9630 IoUM 0.8397 0.8407 0.8494 0.8538 0.8564 Dice 0.7899 0.7994 0.8088 0.8174 0.8230 表 3 在Kvasir-SEG数据集上各实验的量化结果
评估标准 消融实验编号 1 2 3 4 5 召回率 0.8664 0.8631 0.8636 0.8750 0.8752 特异性 0.9840 0.9854 0.9844 0.9858 0.9866 精确率 0.8921 0.9006 0.9163 0.9021 0.9207 F1 0.8560 0.8607 0.8654 0.8689 0.8751 F2 0.8574 0.8673 0.8602 0.8681 0.8718 IoU 0.7866 0.7920 0.7957 0.8032 0.8173 IoUB 0.9534 0.9539 0.9520 0.9532 0.9577 IoUM 0.8700 0.8730 0.8738 0.8782 0.8875 Dice 0.8560 0.8607 0.8654 0.8689 0.8751 表 4 不同模型在EndoSceneStill数据集中的量化评估结果
模型 召回率 特异性 精确率 F1 F2 IoU IoUB IoUM Dice U-Net 0.6839 0.9954 0.9222 0.7113 0.6910 0.6314 0.9515 0.7914 0.7113 Attention unet 0.6744 0.9962 0.9373 0.7084 0.6833 0.6260 0.9504 0.7882 0.7084 TKCN 0.8110 0.9866 0.8565 0.7819 0.7875 0.7023 0.9536 0.8280 0.7819 Xception 0.8017 0.9920 0.8964 0.7940 0.7906 0.7220 0.9575 0.8398 0.7940 DeepLabV3+ 0.7611 0.9919 0.8543 0.7542 0.7505 0.6833 0.9545 0.8189 0.7542 PraNet 0.7973 0.9937 0.9215 0.8016 0.7945 0.7349 0.9610 0.8480 0.8016 SU-Net 0.8237 0.9929 0.9007 0.8230 0.8175 0.7499 0.9630 0.8564 0.8230 表 5 不同模型在Kvasir-SEG数据集中的量化评估结果
模型 召回率 特异性 精确率 F1 F2 IoU IoUB IoUM Dice U-Net 0.8408 0.9707 0.8315 0.8017 0.8161 0.7099 0.9331 0.8215 0.8017 Attention unet 0.8576 0.9682 0.8317 0.8105 0.8283 0.7249 0.9340 0.8294 0.8105 TKCN 0.8651 0.9826 0.8989 0.8552 0.8567 0.7811 0.9473 0.8642 0.8552 Xception 0.8702 0.9831 0.9041 0.8662 0.8639 0.7982 0.9504 0.8743 0.8662 DeepLabV3+ 0.8879 0.9812 0.8938 0.8725 0.8770 0.8110 0.9550 0.8830 0.8725 PraNet 0.8763 0.9859 0.9154 0.8743 0.8718 0.8110 0.9557 0.8833 0.8743 SU-Net 0.8752 0.9866 0.9207 0.8751 0.8718 0.8173 0.9577 0.8875 0.8751 -
[1] GSCHWANTLER M, KRIWANEK S, LANGNER E, et al. High-grade dysplasia and invasive carcinoma in colorectal adenomas: A multivariate analysis of the impact of adenoma and patient characteristics[J]. European Journal of Gastroenterology & Hepatology, 2002, 14(2): 183–188. doi: 10.1097/00042737-200202000-00013 [2] ARNOLD M, SIERRA M S, LAVERSANNE M, et al. Global patterns and trends in colorectal cancer incidence and mortality[J]. Gut, 2017, 66(4): 683–691. doi: 10.1136/gutjnl-2015-310912 [3] PUYAL J G B, BHATIA K K, BRANDAO P, et al. Endoscopic polyp segmentation using a hybrid 2D/3D CNN[C]. 23rd International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 2020: 295–305. [4] TASHK A, HERP J, and NADIMI E. Fully automatic polyp detection based on a novel u-net architecture and morphological post-process[C]. 2019 IEEE International Conference on Control, Artificial Intelligence, Robotics & Optimization, Athens, Greece, 2019: 37–41. [5] WANG Pu, XIAO Xiao, BROWN J R G, et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy[J]. Nature Biomedical Engineering, 2018, 2(10): 741–748. doi: 10.1038/s41551-018-0301-3 [6] SORNAPUDI S, MENG F, and YI S. Region-based automated localization of colonoscopy and wireless capsule endoscopy polyps[J]. Applied Sciences, 2019, 9(12): 2404. doi: 10.3390/app9122404 [7] FAN Dengping, JI Gepeng, ZHOU Tao, et al. PraNet: Parallel reverse attention network for polyp segmentation[C]. 23rd International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 2020: 263–273. [8] FENG Ruiwei, LEI Biwen, WANG Wenzhe, et al. SSN: A stair-shape network for real-time polyp segmentation in colonoscopy images[C]. 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, USA, 2020: 225–229. [9] JI Gepeng, CHOU Yucheng, FAN Dengping, et al. Progressively normalized self-attention network for video polyp segmentation[J]. arXiv: 2105.08468, 2021. [10] LIN Ailiang, CHEN Bingzhi, XU Jiayu, et al. DS-TransUNet: Dual swin transformer U-Net for medical image segmentation[J]. arXiv: 2106.06716, 2021. [11] ZHANG Yundong, LIU Huiye, and HU Qiang. TransFuse: Fusing transformers and CNNs for medical image segmentation[J]. arXiv: 2102.08005, 2021. [12] JHA D, SMEDSRUD P H, RIEGLER M A, et al. Kvasir-SEG: A segmented polyp dataset[C]. 26th International Conference on Multimedia Modeling, Daejeon, Korea, 2020: 451–462. [13] VÁZQUEZ D, BERNAL J, SÁNCHEZ F J, et al. A benchmark for endoluminal scene segmentation of colonoscopy images[J]. Journal of Healthcare Engineering, 2017, 2017: 4037190. doi: 10.1155/2017/4037190 [14] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 2015: 234–241. [15] LONG J, SHELHAMER E, and DARRELL T. Fully convolutional networks for semantic segmentation[C]. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, USA, 2015: 3431–3440. [16] WU Tianyi, TANG Sheng, ZHANG Rui, et al. Tree-structured kronecker convolutional network for semantic segmentation[C]. 2019 IEEE International Conference on Multimedia and Expo (ICME), Shanghai, China, 2019: 940–945. [17] CHOLLET F. Xception: Deep learning with depthwise separable convolutions[C]. The 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, USA, 2017: 1800–1807. [18] HE Kaiming, ZHANG Xiangyu, REN Shaoqing, et al. Deep residual learning for image recognition[C]. The 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, USA, 2016: 770–778. [19] SHI Wenzhe, CABALLERO J, HUSZÁR F, et al. Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network[C]. The 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, USA, 2016: 1874–1883. [20] KINGMA D P and BA J. Adam: A method for stochastic optimization[J]. arXiv: 1412.6980, 2017. [21] OKTAY O, SCHLEMPER J, LE FOLGOC L, et al. Attention U-Net: Learning where to look for the pancreas[J]. arXiv: 1804.03999v3, 2018. [22] CHEN L C, ZHU Yukun, PAPANDREOU G, et al. Encoder-decoder with Atrous separable convolution for semantic image segmentation[C]. The 15th European Conference on Computer Vision (ECCV), Munich, Germany, 2018: 833–851. -





 下载:
下载:


 下载:
下载:
