Malignancy Grading of Lung Nodules Based on CT Signs Quantization Analysis
-
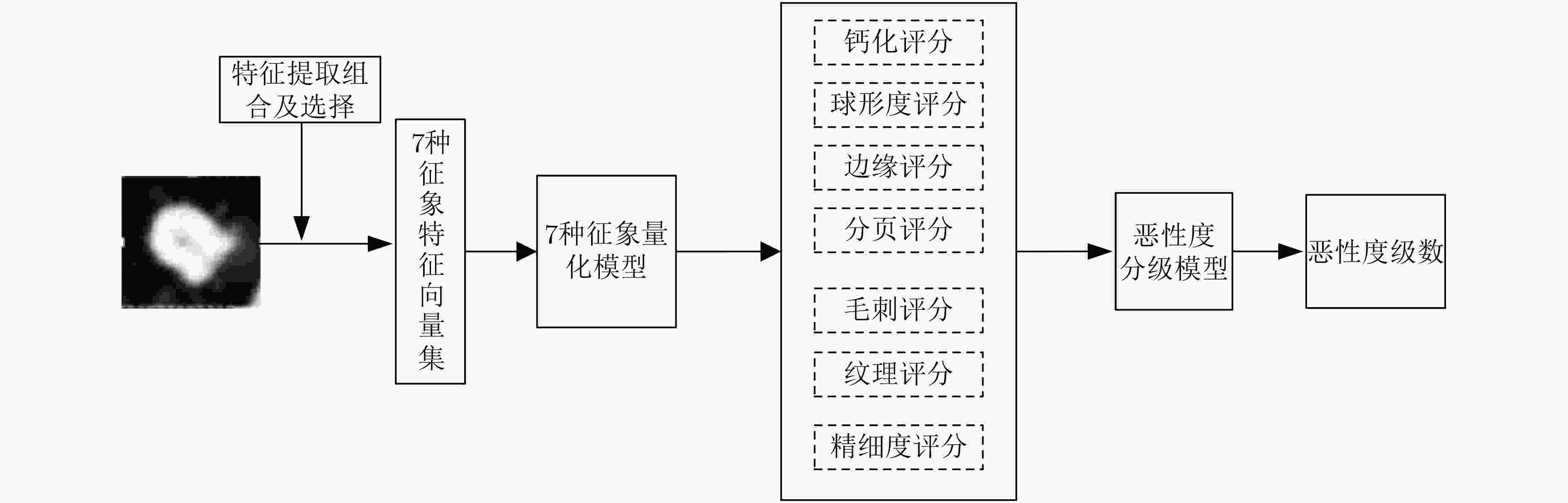
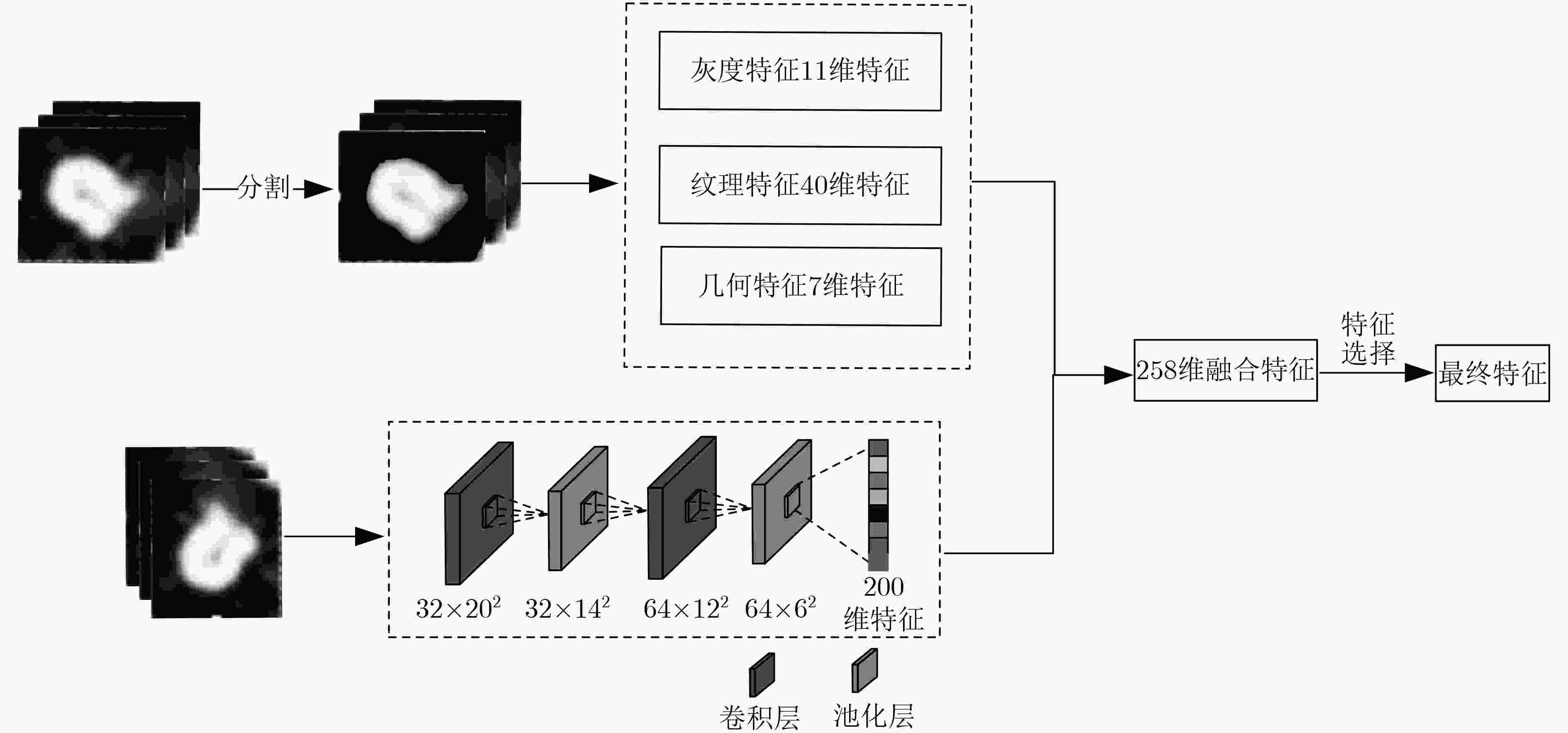
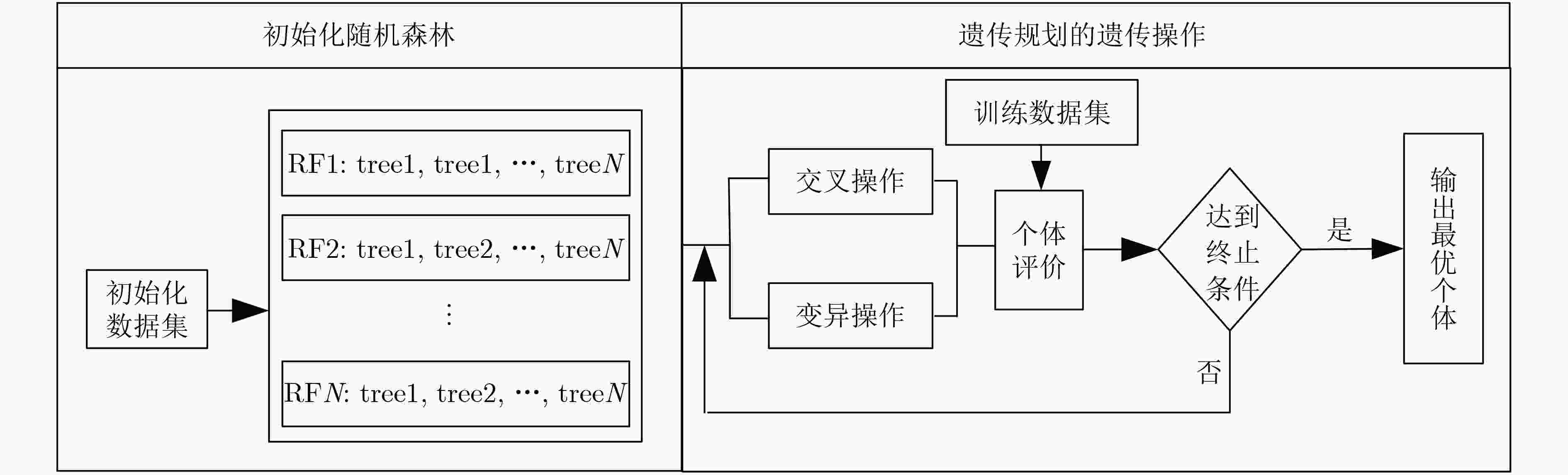
摘要: 为了提高肺结节恶性度分级的计算精度及可解释性,该文提出一种基于CT征象量化分析的肺结节恶性度分级方法。首先,融合影像组学特征和通过卷积神经网络提取的高阶特征构造分析CT征象所需的特征集; 接着,在混合特征集的基础上利用进化搜索机制优化集成学习分类器,实现对7种肺结节征象的识别和量化打分; 最后,将7种CT征象的量化打分输入到一个利用差分进化算法优化产生的多分类器,实现肺结节恶性度的分级计算。在实验研究中使用LIDC-IDRI数据集中的2000个肺结节样本进行进化集成学习器和恶性度分级器的训练和测试。实验结果显示对7种CT征象的识别准确率可达0.9642以上,肺结节恶性度分级的准确率为0.8618,精确率为0.8678,召回率为0.8617,F1指标为0.8627。与多个典型算法的比较显示,该文方法不但具有较高的准确率,而且可对相关CT征象进行量化分析,使得对恶性度的分级结果更具可解释性。Abstract: In order to improve the accuracy and interpretability of the grading of malignant nodules in the lung, a method is proposed to achieve grading automatically for lung nodules by using (Computed Tomography, CT) signs. Firstly, features sets are extracted of CT signs by combing the radiomics features with the higher-order features extracted by convolutional neural network. Then, the ensemble classifier is optimized by the evolutionary search mechanism based on the mixed feature sets, and it is used to realize quantitative scores for 7 CT signs. Finally, 7 quantitative scores are input to the optimized multi-classifier to achieve the grading of malignant nodules in the lung. In the experience, 2000 samples of lung nodules in LIDC-IDRI data set are used to train and test the proposed method. The results show that the recognition accuracy of the 7 CT signs can reach more than 0.9642, the grading accuracy reaches 0.8618, the precision reaches 0.8678, the recall reaches 0.8617, and the F1 index reaches 0.8627. With respect to typical algorithms, the proposed method not only has high accuracy, but also can quantitatively analyze the CT signs that make the grade result of malignancy more interpretive.
-
表 1 不同征象的有效特征
特征 阈值 特征总数量 影像组学特征数 CNN特征数 精细度 0.0050 69 28 41 球形度 0.0030 136 23 113 边缘 0.0035 100 28 72 分叶征 0.0009 203 34 169 毛刺征 0.0060 55 23 22 纹理征 0.0070 39 12 27 钙化征 0.0300 68 20 48 表 2 恶性度分级和CT征象量化的精度
指标 精细度 球形度 边缘 分叶征 毛刺征 纹理征 钙化征 恶性度 ACC 0.9668 0.9764 0.9642 0.9693 0.9792 0.9728 0.9844 0.8618 Pre 0.9648 0.9772 0.9659 0.9522 0.9695 0.9733 0.9683 0.8678 Rec 0.9651 0.9785 0.9346 0.9497 0.9708 0.9736 0.9674 0.8619 F1 0.9649 0.9778 0.9499 0.9509 0.9701 0.9735 0.9678 0.8627 表 3 恶性度分级模型的权重系数
恶性度等级 精细度 球形度 边缘 分叶征 毛刺征 纹理征 钙化征 1 –0.0703 –0.8560 0.3835 0.2935 0.2751 0.0848 –0.7668 2 –0.0452 0.3054 0.5043 –0.0062 0.0069 –0.6435 –0.4618 3 –0.4187 –0.3507 0.4151 0.3337 0.17592 –0.0367 0.5758 4 0.1984 –0.1948 0.4221 0.0369 0.3217 0.0113 0.2751 5 0.8548 –0.4759 0.2756 –0.2990 0.4641 –0.1176 0.5337 表 4 不同集成学习器的量化计算结果对比
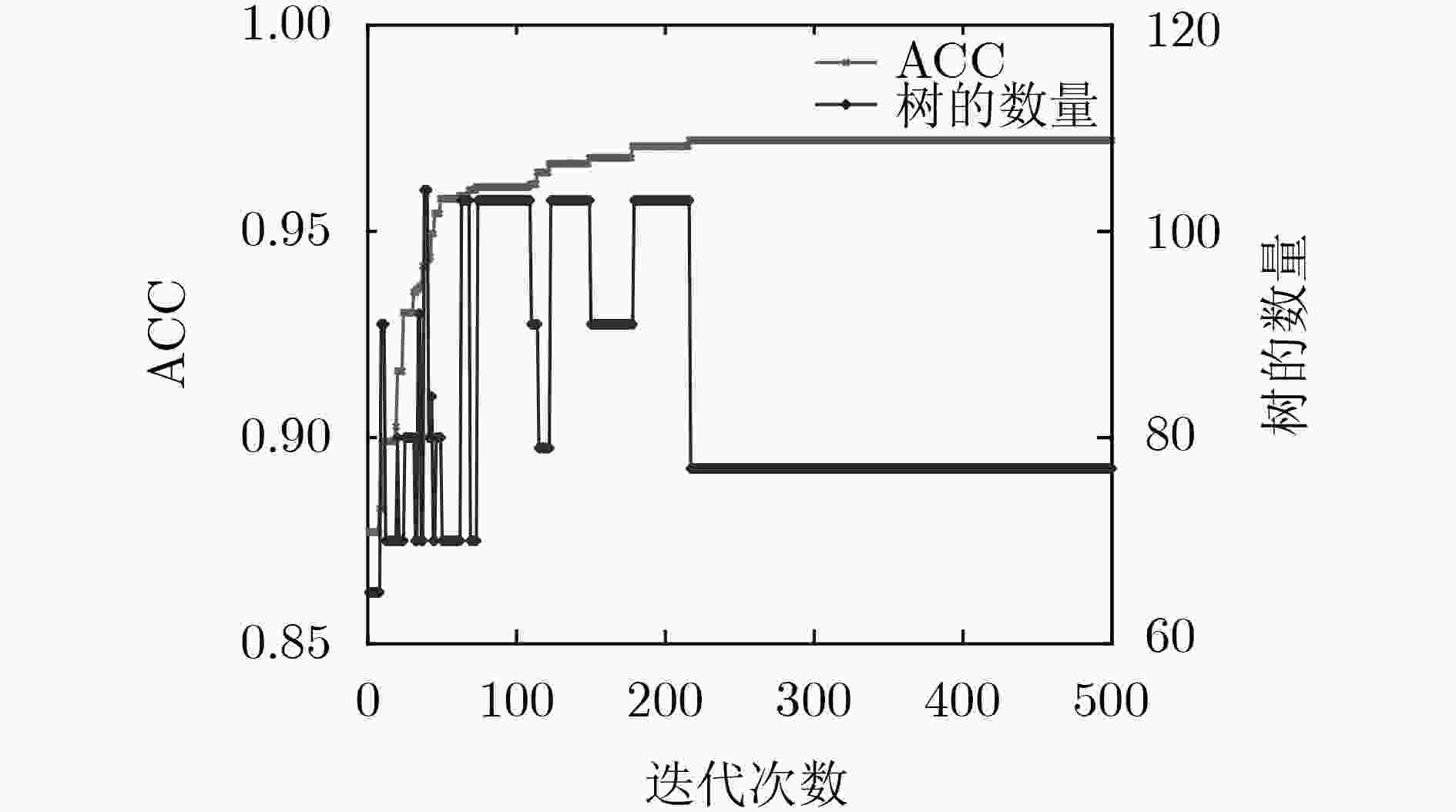
对比分类器 指标 精细度 球形度 边缘 分叶征 毛刺征 纹理征 钙化征 ET ACC 0.9638 0.9526 0.9422 0.9603 0.9372 0.9572 0.9512 Pre 0.9637 0.9558 0.9506 0.9616 0.9372 0.9574 0.9507 Rec 0.9646 0.9532 0.8994 0.9590 0.9374 0.9572 0.9517 F1 0.9638 0.9541 0.9197 0.9598 0.9365 0.9573 0.9511 树个数 112 100 88 108 152 76 76 XGBoost ACC 0.9621 0.9428 0.9452 0.9560 0.9275 0.9312 0.9498 Pre 0.9619 0.9442 0.9504 0.9571 0.9276 0.9311 0.9489 Rec 0.9625 0.9437 0.9266 0.9566 0.9289 0.9311 0.9506 F1 0.9621 0.9438 0.9377 0.9566 0.9282 0.9311 0.9496 树个数 188 180 192 188 176 110 80 RF ACC 0.9585 0.9411 0.9422 0.9491 0.9490 0.9637 0.9471 Pre 0.9583 0.9449 0.9399 0.9497 0.9492 0.9642 0.9466 Rec 0.9593 0.9422 0.9166 0.9488 0.9502 0.9637 0.9477 F1 0.9586 0.9432 0.9270 0.9491 0.9488 0.9639 0.9470 树个数 128 172 188 136 128 128 72 本文方法 ACC 0.9668 0.9764 0.9642 0.9693 0.9792 0.9728 0.9844 Pre 0.9648 0.9772 0.9659 0.9522 0.9695 0.9733 0.9683 Rec 0.9651 0.9785 0.9346 0.9697 0.9708 0.9736 0.9674 F1 0.9649 0.9778 0.9499 0.9509 0.9701 0.9735 0.9678 树个数 77 67 78 54 70 60 23 表 5 相关文献的量化结果对比
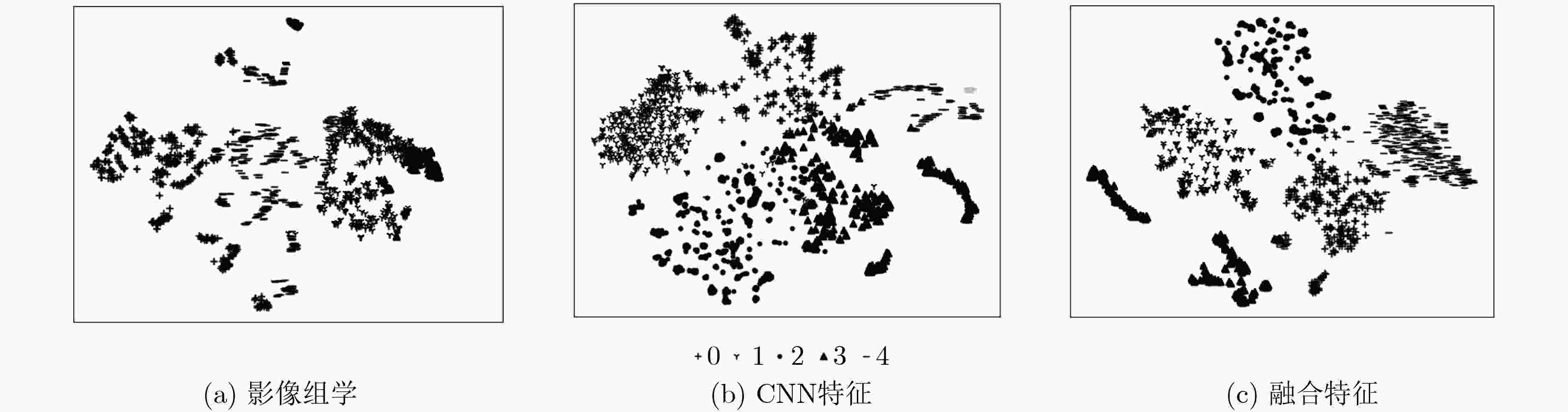
表 6 不同特征集合的聚类结果对比
特征集 均一性 v-measure 互信息 影像组学特征 0.32610 0.3233 0.3179 CNN特征 0.44342 0.4282 0.4118 融合特征 0.60850 0.5934 0.5771 -
[1] MCWILLIAMS A, TAMMEMAGI M C, MAYO J R, et al. Probability of cancer in pulmonary nodules detected on first screening CT[J]. New England Journal of Medicine, 2013, 369(10): 910–919. doi: 10.1056/NEJMoa1214726 [2] NAIDICH D P, BANKIER A A, MACMAHON H, et al. Recommendations for the management of subsolid pulmonary nodules detected at CT: A statement from the fleischner society[J]. Radiology, 2013, 266(1): 304–317. doi: 10.1148/radiol.12120628 [3] GOULD M K, DONINGTON J, LYNCH W R, et al. Evaluation of individuals with pulmonary nodules: When is it lung cancer?: Diagnosis and management of lung cancer, 3rd ed: American college of chest physicians evidence-based clinical practice guidelines[J]. Chest, 2013, 143(5S): e93S–e120S. doi: 10.1378/chest.12-2351 [4] TRAVIS W D, BRAMBILLA E, NOGUCHI M, et al. International association for the study of lung cancer/American thoracic society/European respiratory society: International multidisciplinary classification of lung adenocarcinoma[J]. Proceedings of the American Thoracic Society, 2011, 8(5): 381–385. doi: 10.1513/pats.201107-042ST [5] RODRIGUES M B, DA NÓBREGA R V M, ALVES S S A, et al. Health of things algorithms for malignancy level classification of lung nodules[J]. IEEE Access, 2018, 6: 18592–18601. doi: 10.1109/ACCESS.2018.2817614 [6] DA NÓBREGA R V M, PEIXOTO S A, DA SILVA S S P, et al. Lung nodule classification via deep transfer learning in CT lung images[C]. The IEEE 31st International Symposium on Computer-Based Medical Systems, Karlstad, Sweden, 2018: 244–249. [7] ZUO Wangxia, ZHOU Fuqiang, LI Zuoxin, et al. Multi-resolution CNN and knowledge transfer for candidate classification in lung nodule detection[J]. IEEE Access, 2019, 7: 32510–32521. doi: 10.1109/ACCESS.2019.2903587 [8] SHEN Shiwen, HAN S X, ABERLE D R, et al. An interpretable deep hierarchical semantic convolutional neural network for lung nodule malignancy classification[J]. Expert Systems with Applications, 2019, 128: 84–95. doi: 10.1016/j.eswa.2019.01.048 [9] WANG Huafeng, ZHAO Tingting, LI Lihong, et al. A hybrid CNN feature model for pulmonary nodule malignancy risk differentiation[J]. Journal of X-Ray Science and Technology, 2018, 26(2): 171–187. doi: 10.3233/XST-17302 [10] 褚征, 于炯. 基于随机森林的流处理检查点性能预测[J]. 电子与信息学报, 2020, 42(6): 1452–1459. doi: 10.11999/JEIT190552CHU Zheng and YU Jiong. Performance prediction based on random forest for the stream processing checkpoint[J]. Journal of Electronics &Information Technology, 2020, 42(6): 1452–1459. doi: 10.11999/JEIT190552 [11] NISHIO M, NISHIZAWA M, SUGIYAMA O, et al. Computer-aided diagnosis of lung nodule using gradient tree boosting and bayesian optimization[J]. PLoS One, 2018, 13(4): e0195875. doi: 10.1371/journal.pone.0195875 [12] 吴艇帆, 张仁寿. 机器学习在肺内恶性磨玻璃密度结节的应用研究[J]. 广州大学学报: 自然科学版, 2018, 17(3): 33–39.WU Tingfan and ZHANG Renshou. Research on the application of machine learning in the malignant grinding glass density nodules of lung[J]. Journal of Guangzhou University:Natural Science Edition, 2018, 17(3): 33–39. [13] FERREIRA JR J R, OLIVEIRA M C, and DE AZEVEDO-MARQUES P M. Characterization of pulmonary nodules based on features of margin sharpness and texture[J]. Journal of Digital Imaging, 2018, 31(4): 451–463. doi: 10.1007/s10278-017-0029-8 [14] WU Wenhao, HU Huihui, GONG Jing, et al. Malignant-benign classification of pulmonary nodules based on random forest aided by clustering analysis[J]. Physics in Medicine & Biology, 2019, 64(3): 035017. doi: 10.1088/1361-6560/aafab0 [15] SHAUKAT F, RAJA G, ASHRAF R, et al. Artificial neural network based classification of lung nodules in CT images using intensity, shape and texture features[J]. Journal of Ambient Intelligence and Humanized Computing, 2019, 10(10): 4135–4159. doi: 10.1007/s12652-019-01173-w [16] 李双双, 侯震, 刘娟, 等. 影像组学分析与建模工具综述[J]. 中国医学物理学杂志, 2018, 35(9): 1043–1049. doi: 10.3969/j.issn.1005-202X.2018.09.010LI Shuangshuang, HOU Zhen, LIU Juan, et al. Review of radiomic analysis and modeling tools[J]. Chinese Journal of Medical Physics, 2018, 35(9): 1043–1049. doi: 10.3969/j.issn.1005-202X.2018.09.010 [17] SHI Zhenghao, HAO Huan, ZHAO Minghua, et al. A deep CNN based transfer learning method for false positive reduction[J]. Multimedia Tools and Applications, 2019, 78(1): 1017–1033. doi: 10.1007/s11042-018-6082-6 [18] 刘家辰, 苗启广, 曹莹, 等. 基于混合多样性生成与修剪的集成单类分类算法[J]. 电子与信息学报, 2015, 37(2): 386–393. doi: 10.11999/JEIT140161LIU Jiachen, MIAO Qiguang, CAO Ying, et al. Ensemble one-class classifiers based on hybrid diversity generation and pruning[J]. Journal of Electronics &Information Technology, 2015, 37(2): 386–393. doi: 10.11999/JEIT140161 [19] ELYAN E and GABER M M. A fine-grained random forests using class decomposition: An application to medical diagnosis[J]. Neural Computing and Applications, 2016, 27(8): 2279–2288. doi: 10.1007/s00521-015-2064-z [20] MIAO Fen, CAI Yunpeng, ZHANG Yuxiao, et al. Predictive modeling of hospital mortality for patients with heart failure by using an improved random survival forest[J]. IEEE Access, 2018, 6: 7244–7253. doi: 10.1109/ACCESS.2018.2789898 [21] PAUL A, MUKHERJEE D P, DAS P, et al. Improved random forest for classification[J]. IEEE Transactions on Image Processing, 2018, 27(8): 4012–4024. doi: 10.1109/tip.2018.2834830 [22] WANG Qingfeng, CHENG Jiezhi, LIU Zhiqin, et al. Multi-order transfer learning for pathologic diagnosis of pulmonary nodule malignancy[C]. 2018 IEEE International Conference on Bioinformatics and Biomedicine, Madrid, Spain, 2018: 2813–2815. [23] YANG Jing, LI Na, FANG Shuai, et al. Semantic features prediction for pulmonary nodule diagnosis based on online streaming feature selection[J]. IEEE Access, 2019, 7: 61121–61135. doi: 10.1109/ACCESS.2019.2903682 [24] WU Botong, ZHOU Zhen, WANG Jianwei, et al. Joint learning for pulmonary nodule segmentation, attributes and malignancy prediction[C]. The IEEE 15th International Symposium on Biomedical Imaging, Washington, USA, 2018: 1109–1113. -





 下载:
下载:


 下载:
下载:
