Joint Mask and Multi-Frequency Dual Attention GAN Network for CT-to-DWI Image Synthesis in Acute Ischemic Stroke
-
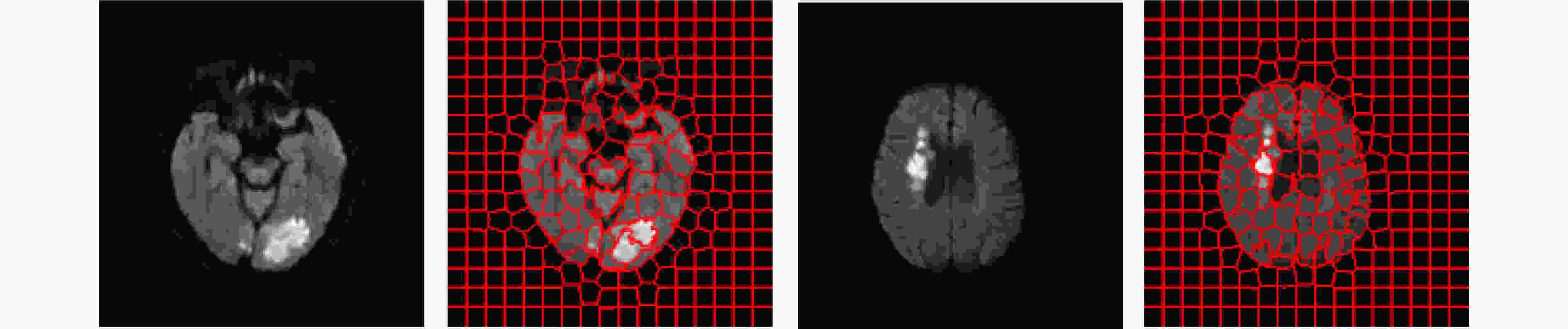
摘要: 基于人工智能的跨模态医学图像生成技术为急性缺血性脑卒中的快速多模态诊疗提供了新的路径。针对现有医学图像生成方法仅依赖图像数据本身的统计特征、忽略医学图像的解剖结构,从而造成病灶模糊和结构偏差问题,该文提出了一种新的联合掩码与多频双重注意力GAN模型,用于急性脑缺血性卒中CT到DWI影像生成。该模型主要包含:(1)掩码引导特征融合模块:通过CT图像与掩码图像的卷积融合,引入解剖结构的空间先验信息,增强脑区及病灶区域的特征表达;(2)多频域注意力编码器:采用离散小波变换分解低频全局特征与高频边缘特征,通过双通路注意力跨尺度融合,减少深层信息的丢失;(3)自适应融合权重模块:结合卷积神经网络与注意力机制,自动学习每个输入特征的自适应权重系数。该研究在临床CT到DWI多模态急性脑缺血性卒中数据集上开展了实验验证,分别在全局尺度采用均方误差、峰值信噪比、结构相似度指数进行评估,在局部尺度基于超像素分割后统计灰度均值相关性进行分析。结果表明,所提模型在各项指标上均优于当前先进方法,对脑区轮廓和病灶区域具有更高的准确性和还原性。Abstract:
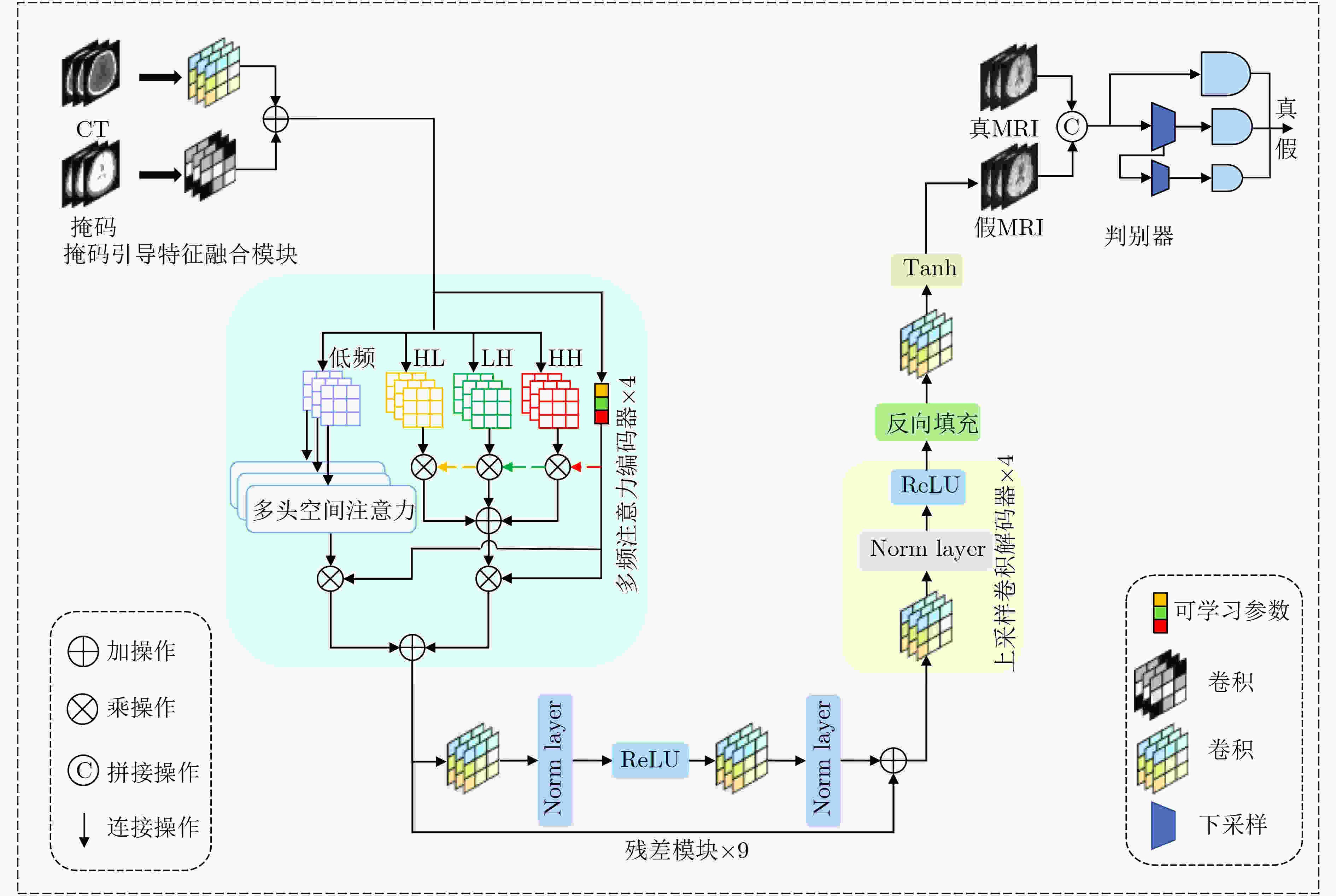
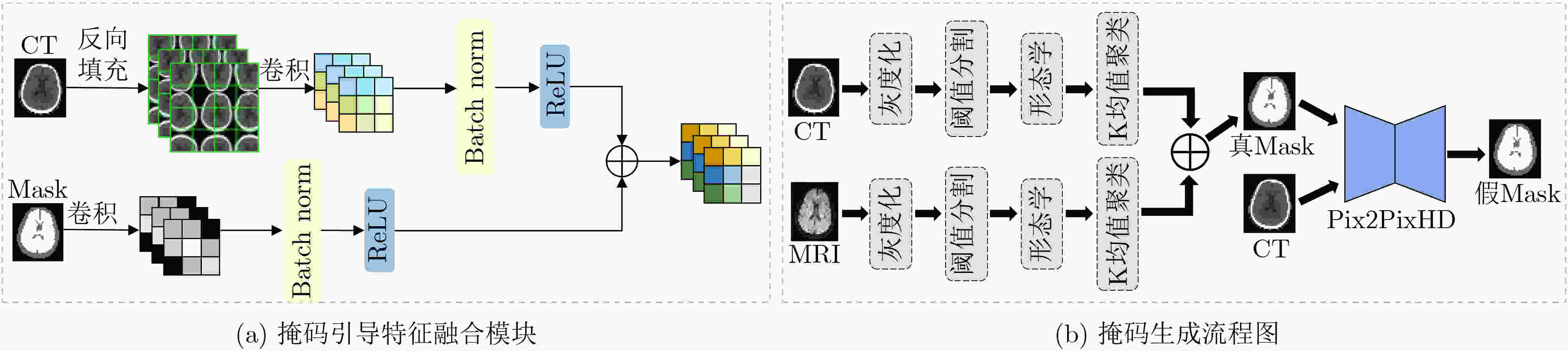
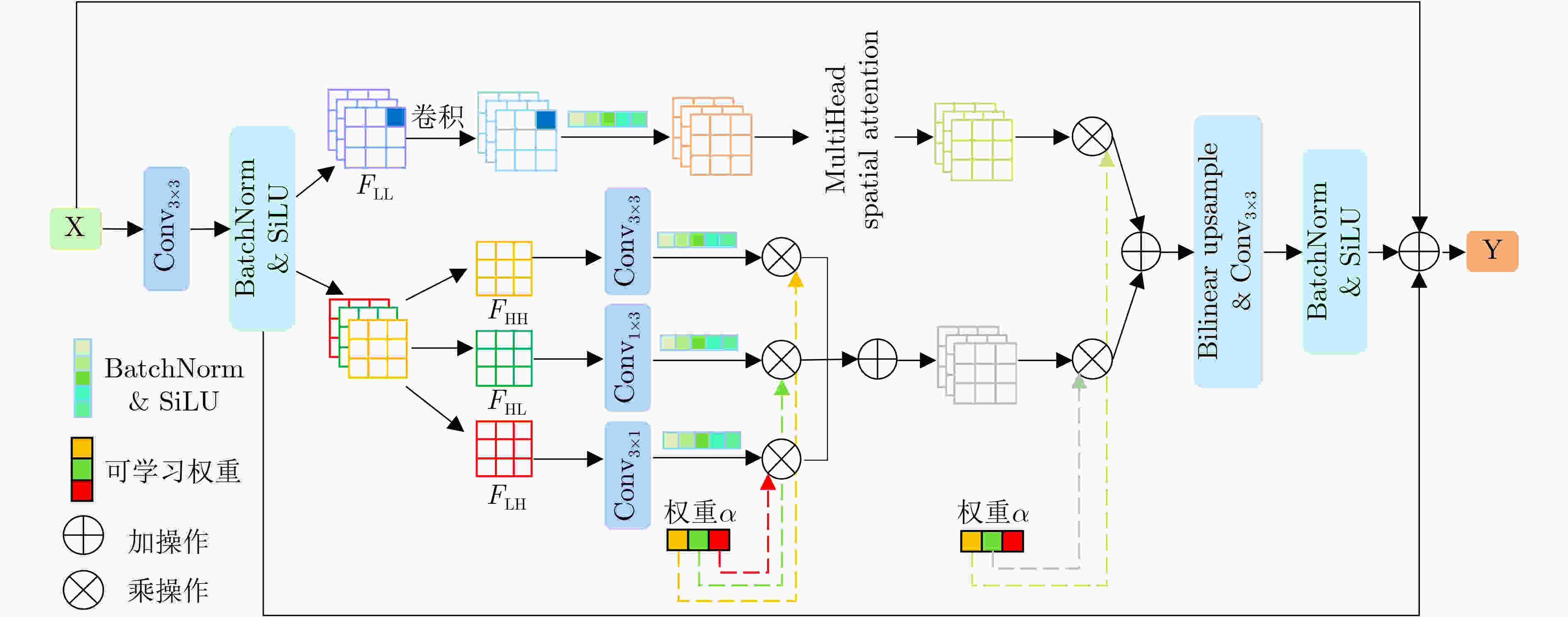
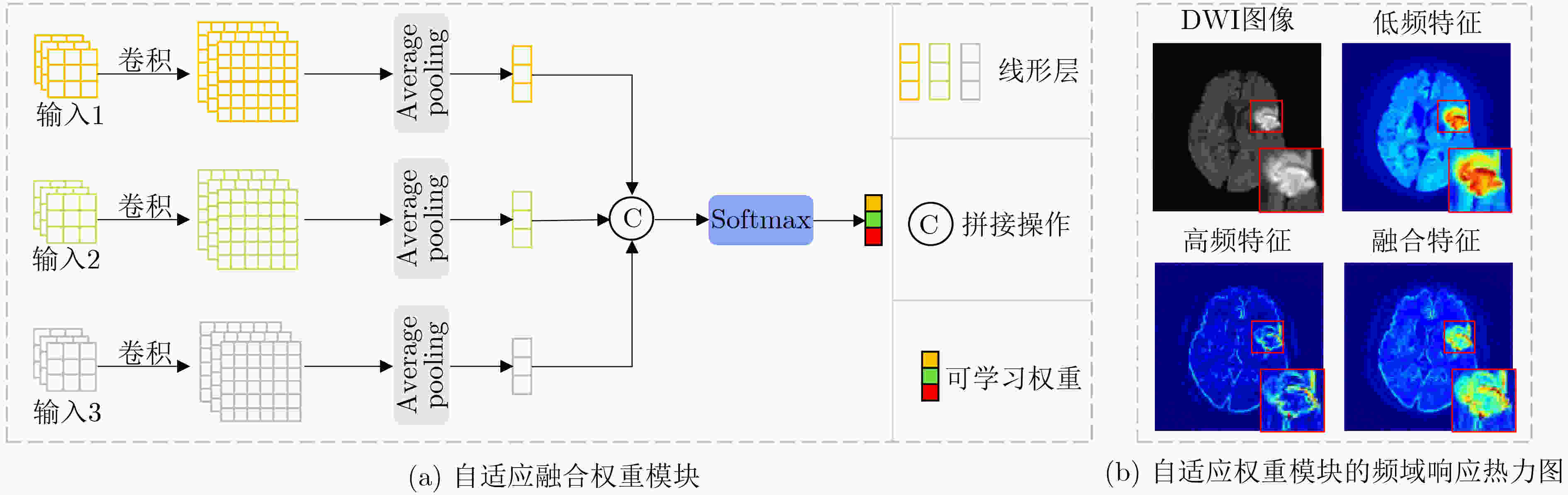
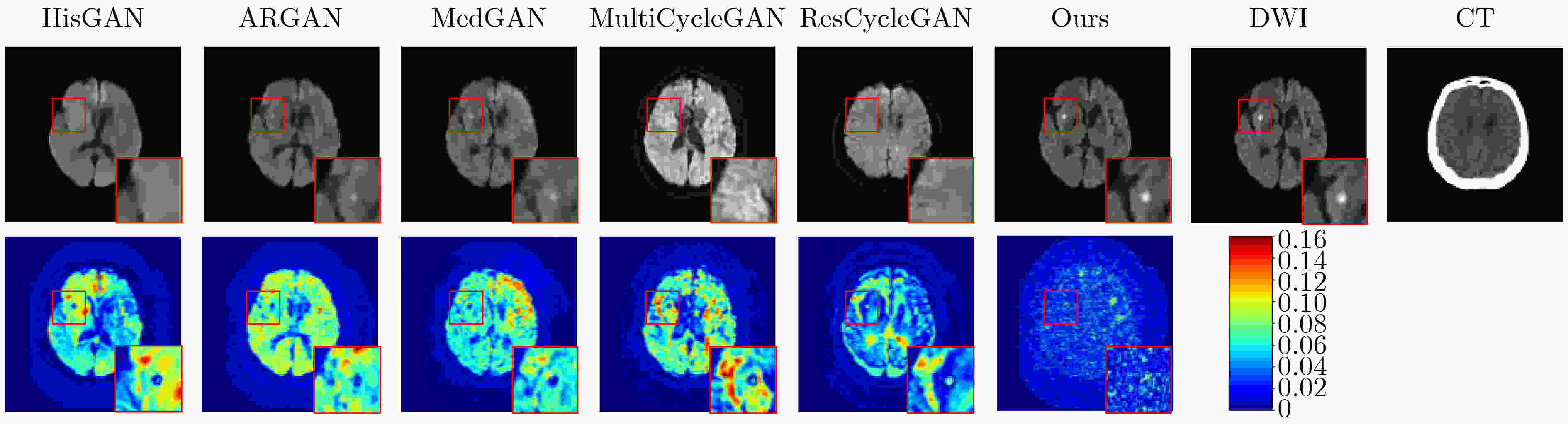
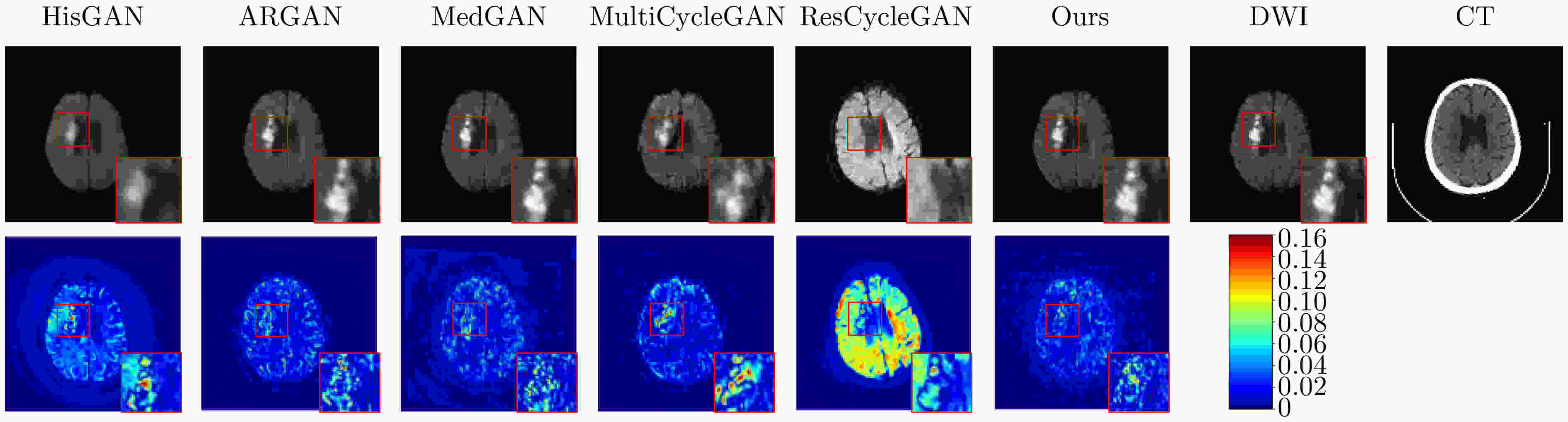
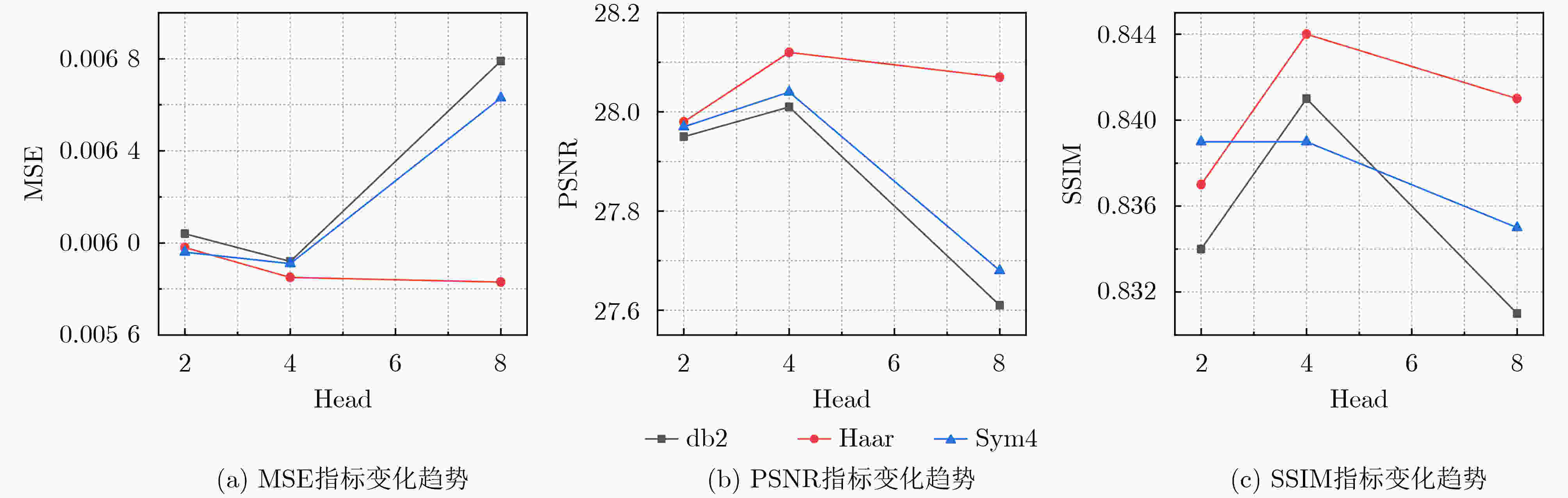
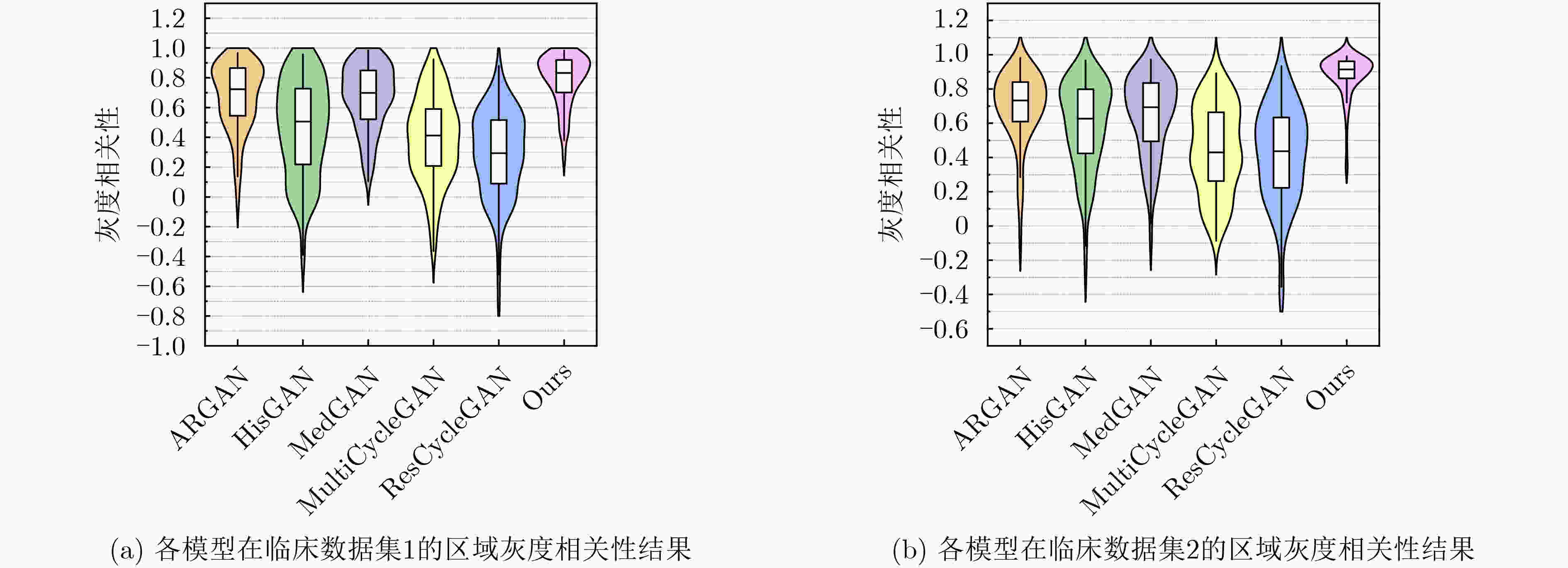
Objective In the clinical management of Acute Ischemic Stroke (AIS), Computed Tomography (CT) and Diffusion-Weighted Imaging (DWI) serve complementary roles at different stages. CT is widely applied for initial evaluation due to its rapid acquisition and accessibility, but it has limited sensitivity in detecting early ischemic changes, which can result in diagnostic uncertainty. In contrast, DWI demonstrates high sensitivity to early ischemic lesions, enabling visualization of diffusion-restricted regions soon after symptom onset. However, DWI acquisition requires a longer time, is susceptible to motion artifacts, and depends on scanner availability and patient cooperation, thereby reducing its clinical accessibility. The limited availability of multimodal imaging data remains a major challenge for timely and accurate AIS diagnosis. Therefore, developing a method capable of rapidly and accurately generating DWI images from CT scans has important clinical significance for improving diagnostic precision and guiding treatment planning. Existing medical image translation approaches primarily rely on statistical image features and overlook anatomical structures, which leads to blurred lesion regions and reduced structural fidelity. Methods This study proposes a Joint Mask and Multi-Frequency Dual Attention Generative Adversarial Network (JMMDA-GAN) for CT-to-DWI image synthesis to assist in the diagnosis and treatment of ischemic stroke. The approach incorporates anatomical priors from brain masks and adaptive multi-frequency feature fusion to improve image translation accuracy. JMMDA-GAN comprises three principal modules: a mask-guided feature fusion module, a multi-frequency attention encoder, and an adaptive fusion weighting module. The mask-guided feature fusion module integrates CT images with anatomical masks through convolution, embedding spatial priors to enhance feature representation and texture detail within brain regions and ischemic lesions. The multi-frequency attention encoder applies Discrete Wavelet Transform (DWT) to decompose images into low-frequency global components and high-frequency edge components. A dual-path attention mechanism facilitates cross-scale feature fusion, reducing high-frequency information loss and improving structural detail reconstruction. The adaptive fusion weighting module combines convolutional neural networks and attention mechanisms to dynamically learn the relative importance of input features. By assigning adaptive weights to multi-scale features, the module selectively enhances informative regions and suppresses redundant or noisy information. This process enables effective integration of low- and high-frequency features, thereby improving both global contextual consistency and local structural precision. Results and Discussions Extensive experiments were performed on two independent clinical datasets collected from different hospitals to assess the effectiveness of the proposed method. JMMDA-GAN achieved Mean Squared Error (MSE) values of 0.0097 and 0.0059 on Clinical Dataset 1 and Clinical Dataset 2, respectively, exceeding state-of-the-art models by reducing MSE by 35.8% and 35.2% compared with ARGAN. The proposed network reached peak Signal-to-Noise Ratio (PSNR) values of 26.75 and 28.12, showing improvements of 30.7% and 7.9% over the best existing methods. For Structural Similarity Index (SSIM), JMMDA-GAN achieved 0.753 and 0.844, indicating superior structural preservation and perceptual quality. Visual analysis further demonstrates that JMMDA-GAN restores lesion morphology and fine texture features with higher fidelity, producing sharper lesion boundaries and improved structural consistency compared with other methods. Cross-center generalization and multi-center mixed experiments confirm that the model maintains stable performance across institutions, highlighting its robustness and adaptability in clinical settings. Parameter sensitivity analysis shows that the combination of Haar wavelet and four attention heads achieves an optimal balance between global structural retention and local detail reconstruction. Moreover, superpixel-based gray-level correlation experiments demonstrate that JMMDA-GAN exceeds existing models in both local consistency and global image quality, confirming its capacity to generate realistic and diagnostically reliable DWI images from CT inputs. Conclusions This study proposes a novel JMMDA-GAN designed to enhance lesion and texture detail generation by incorporating anatomical structural information. The method achieves this through three principal modules. (1) The mask-guided feature fusion module effectively integrates anatomical structure information, with particular optimization of the lesion region. The mask-guided network focuses on critical lesion features, ensuring accurate restoration of lesion morphology and boundaries. By combining mask and image data, the method preserves the overall anatomical structure while enhancing lesion areas, preventing boundary blurring and texture loss commonly observed in traditional approaches, thereby improving diagnostic reliability. (2) The multi-frequency feature fusion module jointly optimizes low- and high-frequency features to enhance image detail. This integration preserves global structural integrity while refining local features, producing visually realistic and high-fidelity images. (3) The adaptive fusion weighting module dynamically adjusts the learning strategy for frequency-domain features according to image content, enabling the network to manage texture variations and complex anatomical structures effectively, thereby improving overall image quality. Through the coordinated function of these modules, the proposed method enhances image realism and diagnostic precision. Experimental results demonstrate that JMMDA-GAN exceeds existing advanced models across multiple clinical datasets, highlighting its potential to support clinicians in the diagnosis and management of AIS. -
表 1 为评估JMMDA-GAN模型性能使用的两个临床数据集信息
数据集 男性数量 女性数量 平均年龄 训练图像 测试图像 临床数据集1 148 80 58 6222 1556 临床数据集2 365 270 64 6533 1634 表 2 临床数据集1的对比实验指标
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 HisGAN 0.0186 ±0.0124 *2×10–163 22.84±2.52* 7×10–205 0.540±0.112* 9×10–255 ARGAN 0.0151 ±0.0113 *4×10–145 24.02±3.20* 6×10–179 0.752±0.074* 3×10–5 MedGAN 0.0187 ±0.0133 *1×10–193 23.14±3.41* 1×10–228 0.713±0.081* 4×10–125 MultiCycleGAN 0.0240 ±0.0165 *9×10–217 21.74±2.50* 9×10–240 0.636±0.098* 3×10–247 ResCycleGAN 0.0344 ±0.0270 *5×10–232 20.44±2.88* 6×10–245 0.596±0.106* 4×10–254 本文方法 0.0097 ±0.0114 - 26.75±4.32 - 0.753±0.101 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异 表 3 临床数据集2的对比实验指标
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 HisGAN 0.0119 ±0.0076 *7×10–223 24.69±2.34* 6×10–246 0.642±0.116* 1×10–268 ARGAN 0.0091 ±0.0065 *4×10–166 26.06±2.79* 2×10–190 0.825±0.063* 7×10–159 MedGAN 0.0112 ±0.0075 *6×10–217 25.12±2.81* 2×10–240 0.796±0.065* 1×10–225 MultiCycleGAN 0.0131 ±0.0072 *1×10–243 24.12±2.10* 6×10–260 0.744±0.077* 1×10–263 ResCycleGAN 0.0283 ±0.0199 *1×10–260 21.09±2.60* 1×10–265 0.660±0.087* 1×10–268 本文方法 0.0059 ±0.0053 - 28.12±2.85 - 0.844±0.072 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异 表 4 跨中心泛化实验指标(临床数据集1训练、临床数据集2测试)
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 HisGAN 0.0337 ±0.0209 *3×10–137 20.24±2.57* 3×10–174 0.601±0.112* 3×10–106 ARGAN 0.0296 ±0.0170 *1×10–181 20.66±2.25* 7×10–220 0.611±0.089* 2×10–218 MedGAN 0.0313 ±0.0206 *4×10–146 20.54±2.45* 5×10–190 0.625±0.074* 4×10–158 MultiCycleGAN 0.0462 ±0.0273 *7×10–256 18.82±2.44* 3×10–263 0.556±0.085* 9×10–263 ResCycleGAN 0.0345 ±0.0240 *1×10–160 20.19±2.56* 1×10–193 0.618±0.079* 5×10–154 本文方法 0.0155 ±0.0154 - 24.17±3.12 - 0.678±0.073 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异 表 5 跨中心泛化实验指标
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 HisGAN 0.0313 ±0.0211 *1×10–174 20.55±2.43* 1×10–205 0.480±0.115* 4×10–251 ARGAN 0.0313 ±0.0224 *3×10–184 20.66±2.59* 1×10–215 0.615±0.090* 2×10–236 MedGAN 0.0294 ±0.0211 *1×10–170 20.88±2.49* 2×10–205 0.620±0.084* 6×10–225 MultiCycleGAN 0.0356 ±0.0242 *2×10–224 20.02±2.46* 7×10–240 0.578±0.092* 5×10–253 ResCycleGAN 0.0394 ±0.0300 *7×10–210 19.82±2.88* 1×10–224 0.585±0.100* 1×10–238 本文方法 0.0161 ±0.0153 - 23.96±3.10 - 0.681±0.084 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异 表 6 多中心混合数据集的对比实验指标
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 HisGAN 0.0156 ±0.0116 *< 0.001 23.68±2.59* < 0.001 0.592±0.125* < 0.001 ARGAN 0.0121 ±0.0095 *1×10–302 24.95±2.97* < 0.001 0.786±0.075* 3×10–77 MedGAN 0.0153 ±0.0111 *< 0.001 23.91±3.09* < 0.001 0.748±0.079* < 0.001 MultiCycleGAN 0.0255 ±0.0187 *< 0.001 21.50±2.49* < 0.001 0.660±0.103* < 0.001 ResCycleGAN 0.0321 ±0.0240 *< 0.001 20.63±2.74* < 0.001 0.623±0.105* < 0.001 本文方法 0.0077 ±0.0080 - 27.22±3.34 - 0.794±0.098 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异,p<0.001表示p值远小于统计软件或数值精度的下限(如<1×10–300)。 表 7 消融实验指标
方法 MSE↓ MSE p值 PSNR↑ PSNR p值 SSIM↑ SSIM p值 Pix2PixHD 0.00969 ±0.0066 *8×10–191 25.78±3.00* 1×10–206 0.813±0.084* 9×10–239 Pix2PixHD+MGFF 0.00681 ±0.0060 *2×10–55 27.41±2.78* 6×10–69 0.830±0.069* 2×10–168 Pix2PixHD+MFAB 0.00809 ±0.0058 *2×10–137 26.62±3.02* 1×10–153 0.831±0.077* 1×10–143 本文方法 0.00585 ±0.0053 - 28.12±2.85 - 0.844±0.072 - 注:*表示本文方法与其他方法在Wilcoxon符号秩检验中取得显著差异 -
[1] ZHANG Xuting, ZHONG Wansi, XUE Rui, et al. Argatroban in patients with acute ischemic stroke with early neurological deterioration: A randomized clinical trial[J]. JAMA Neurology, 2024, 81(2): 118–125. doi: 10.1001/jamaneurol.2023.5093. [2] VANDE VYVERE T, PISICĂ D, WILMS G, et al. Imaging findings in acute traumatic brain injury: A national institute of neurological disorders and stroke common data element-based pictorial review and analysis of over 4000 admission brain computed tomography scans from the collaborative European NeuroTrauma effectiveness research in traumatic brain injury (CENTER-TBI) study[J]. Journal of Neurotrauma, 2024, 41(19/20): 2248–2297. doi: 10.1089/neu.2023.0553. [3] ELSHERIF S, LEGERE B, MOHAMED A, et al. Beyond conventional imaging: A systematic review and meta-analysis assessing the impact of computed tomography perfusion on ischemic stroke outcomes in the late window[J]. International Journal of Stroke, 2025, 20(3): 278–288. doi: 10.1177/17474930241292915. [4] RAPILLO C M, DUNET V, PISTOCCHI S, et al. Moving from CT to MRI paradigm in acute ischemic stroke: Feasibility, effects on stroke diagnosis and long-term outcomes[J]. Stroke, 2024, 55(5): 1329–1338. doi: 10.1161/strokeaha.123.045154. [5] GHEBREHIWET I, ZAKI N, DAMSEH R, et al. Revolutionizing personalized medicine with generative AI: A systematic review[J]. Artificial Intelligence Review, 2024, 57(5): 128. doi: 10.1007/s10462-024-10768-5. [6] SHURRAB S, GUERRA-MANZANARES A, MAGID A, et al. Multimodal machine learning for stroke prognosis and diagnosis: A systematic review[J]. IEEE Journal of Biomedical and Health Informatics, 2024, 28(11): 6958–6973. doi: 10.1109/jbhi.2024.3448238. [7] ARMANIOUS K, JIANG Chenming, FISCHER M, et al. MedGAN: Medical image translation using GANs[J]. Computerized Medical Imaging and Graphics, 2020, 79: 101684. doi: 10.1016/j.compmedimag.2019.101684. [8] EKANAYAKE M, PAWAR K, HARANDI M, et al. McSTRA: A multi-branch cascaded swin transformer for point spread function-guided robust MRI reconstruction[J]. Computers in Biology and Medicine, 2024, 168: 107775. doi: 10.1016/j.compbiomed.2023.107775. [9] DALMAZ O, YURT M, and ÇUKUR T. ResViT: Residual vision transformers for multimodal medical image synthesis[J]. IEEE Transactions on Medical Imaging, 2022, 41(10): 2598–2614. doi: 10.1109/tmi.2022.3167808. [10] ÖZBEY M, DALMAZ O, DAR S U H, et al. Unsupervised medical image translation with adversarial diffusion models[J]. IEEE Transactions on Medical Imaging, 2023, 42(12): 3524–3539. doi: 10.1109/tmi.2023.3290149. [11] LUO Yu, ZHANG Shaowei, LING Jie, et al. Mask-guided generative adversarial network for MRI-based CT synthesis[J]. Knowledge-Based Systems, 2024, 295: 111799. doi: 10.1016/j.knosys.2024.111799. [12] YANG Linlin, SHANGGUAN Hong, ZHANG Xiong, et al. High-frequency sensitive generative adversarial network for low-dose CT image denoising[J]. IEEE Access, 2020, 8: 930–943. doi: 10.1109/access.2019.2961983. [13] HUTCHINSON E B, AVRAM A V, IRFANOGLU M O, et al. Analysis of the effects of noise, DWI sampling, and value of assumed parameters in diffusion MRI models[J]. Magnetic Resonance in Medicine, 2017, 78(5): 1767–1780. doi: 10.1002/mrm.26575. [14] DAS S and KUNDU M K. NSCT-based multimodal medical image fusion using pulse-coupled neural network and modified spatial frequency[J]. Medical & Biological Engineering & Computing, 2012, 50(10): 1105–1114. doi: 10.1007/s11517-012-0943-3. [15] 周涛, 刘赟璨, 陆惠玲, 等. ResNet及其在医学图像处理领域的应用: 研究进展与挑战[J]. 电子与信息学报, 2022, 44(1): 149–167. doi: 10.11999/JEIT210914.ZHOU Tao, LIU Yuncan, LU Huiling, et al. ResNet and its application to medical image processing: Research progress and challenges[J]. Journal of Electronics & Information Technology, 2022, 44(1): 149–167. doi: 10.11999/JEIT210914. [16] BARRON J T. A general and adaptive robust loss function[C]. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, USA, 2019: 4326–4334. doi: 10.1109/CVPR.2019.00446. [17] LUO Jialin, DAI Peishan, HE Zhuang, et al. Deep learning models for ischemic stroke lesion segmentation in medical images: A survey[J]. Computers in Biology and Medicine, 2024, 175: 108509. doi: 10.1016/j.compbiomed.2024.108509. [18] WANG Tingchun, LIU Mingyu, ZHU Junyan, et al. High-resolution image synthesis and semantic manipulation with conditional GANs[C]. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 8798–8807. doi: 10.1109/CVPR.2018.00917. [19] LIU Rui, DING Xiaoxi, SHAO Yimin, et al. An interpretable multiplication-convolution residual network for equipment fault diagnosis via time–frequency filtering[J]. Advanced Engineering Informatics, 2024, 60: 102421. doi: 10.1016/j.aei.2024.102421. [20] LI Yihao, EL HABIB DAHO M, CONZE P H, et al. A review of deep learning-based information fusion techniques for multimodal medical image classification[J]. Computers in Biology and Medicine, 2024, 177: 108635. doi: 10.1016/j.compbiomed.2024.108635. [21] PENG Yanjun, SUN Jindong, REN Yande, et al. A histogram-driven generative adversarial network for brain MRI to CT synthesis[J]. Knowledge-Based Systems, 2023, 277: 110802. doi: 10.1016/j.knosys.2023.110802. [22] LIU Yanxia, CHEN Anni, SHI Hongyu, et al. CT synthesis from MRI using multi-cycle GAN for head-and-neck radiation therapy[J]. Computerized Medical Imaging and Graphics, 2021, 91: 101953. doi: 10.1016/j.compmedimag.2021.101953. [23] DAI Xianjin, LEI Yang, LIU Yingzi, et al. Intensity non-uniformity correction in MR imaging using residual cycle generative adversarial network[J]. Physics in Medicine & Biology, 2020, 65(21): 215025. doi: 10.1088/1361-6560/abb31f. [24] DING Bin, LONG Chengjiang, ZHANG Ling, et al. ARGAN: Attentive recurrent generative adversarial network for shadow detection and removal[C]. 2019 IEEE/CVF International Conference on Computer Vision, Seoul, South Korea, 2019: 10212–10221. doi: 10.1109/ICCV.2019.01031. -





 下载:
下载:


 下载:
下载:
