Wave-MambaCT: Low-dose CT Artifact Suppression Method Based on Wavelet Mamba
-
摘要: 低剂量CT(LDCT)图像中的伪影和噪声影响疾病的早期诊断和治疗。基于卷积神经网络的去噪方法在远程建模方面能力有限。与Transformer架构的远程建模方法相比,基于Mamba模型在建模时计算复杂度低,然而现有的Mamba模型存在信息丢失或噪声残留的缺点。为此,该文提出一种基于小波Mamba的去噪模型Wave-MambaCT。首先利用小波变换的多尺度分解解耦噪声和低频内容信息。其次,构建残差模块结合状态空间模型的Mamba模块提取高低频带的局部和全局信息,并用无噪的低频特征通过基于注意力的跨频Mamba模块校正并增强同尺度的高频特征,在去除噪声的同时保持更多细节。最后,分阶段采用逆小波变换渐进恢复图像,并设置相应的损失函数提高网络的稳定性。实验结果表明Wave-MambaCT在较低的计算复杂度和参数量下,不仅提高了低剂量CT图像的视觉效果,而且在PSNR,SSIM,VIF和MSE 4种定量指标上均优于现有的去噪方法。Abstract:
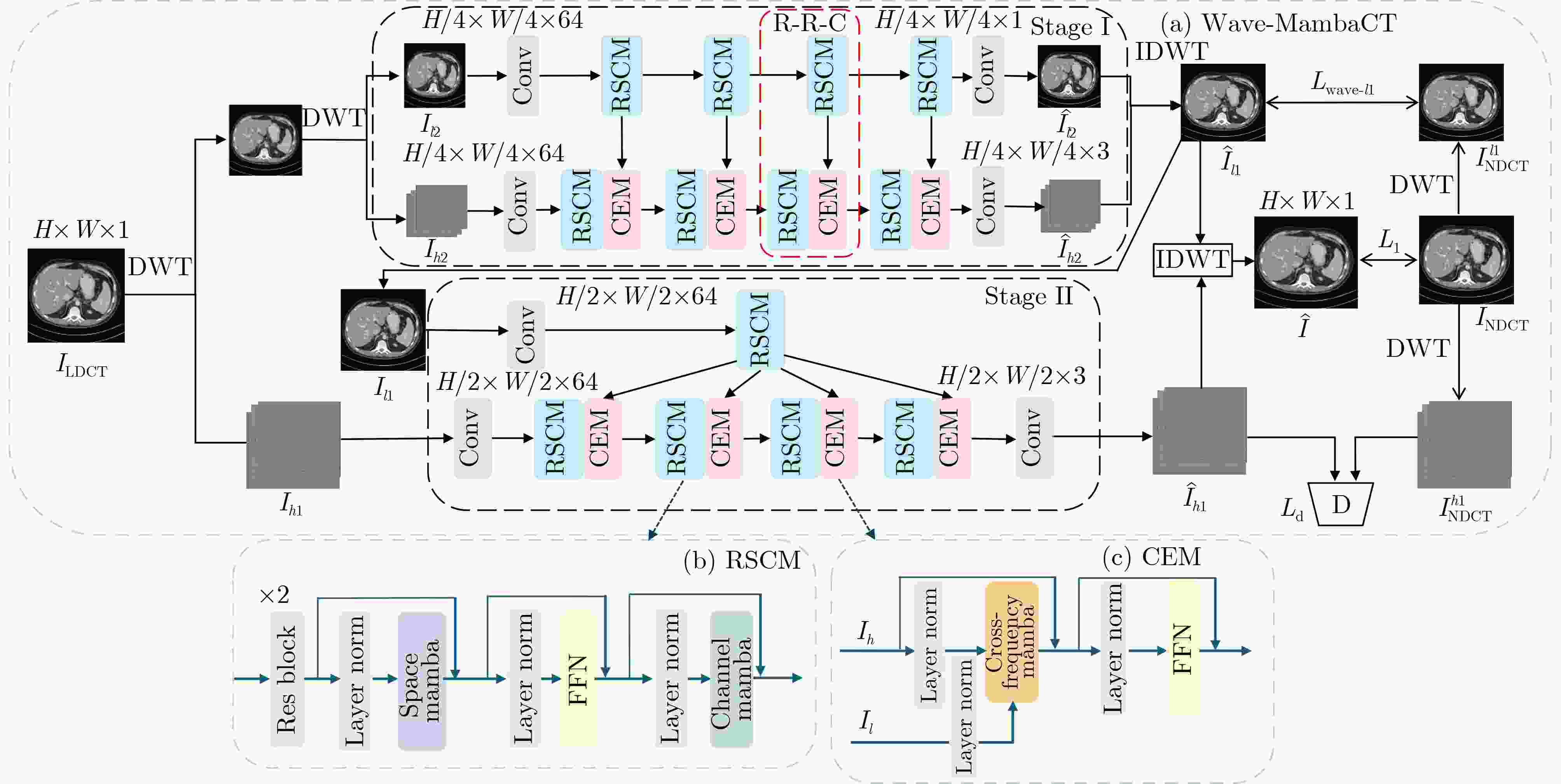
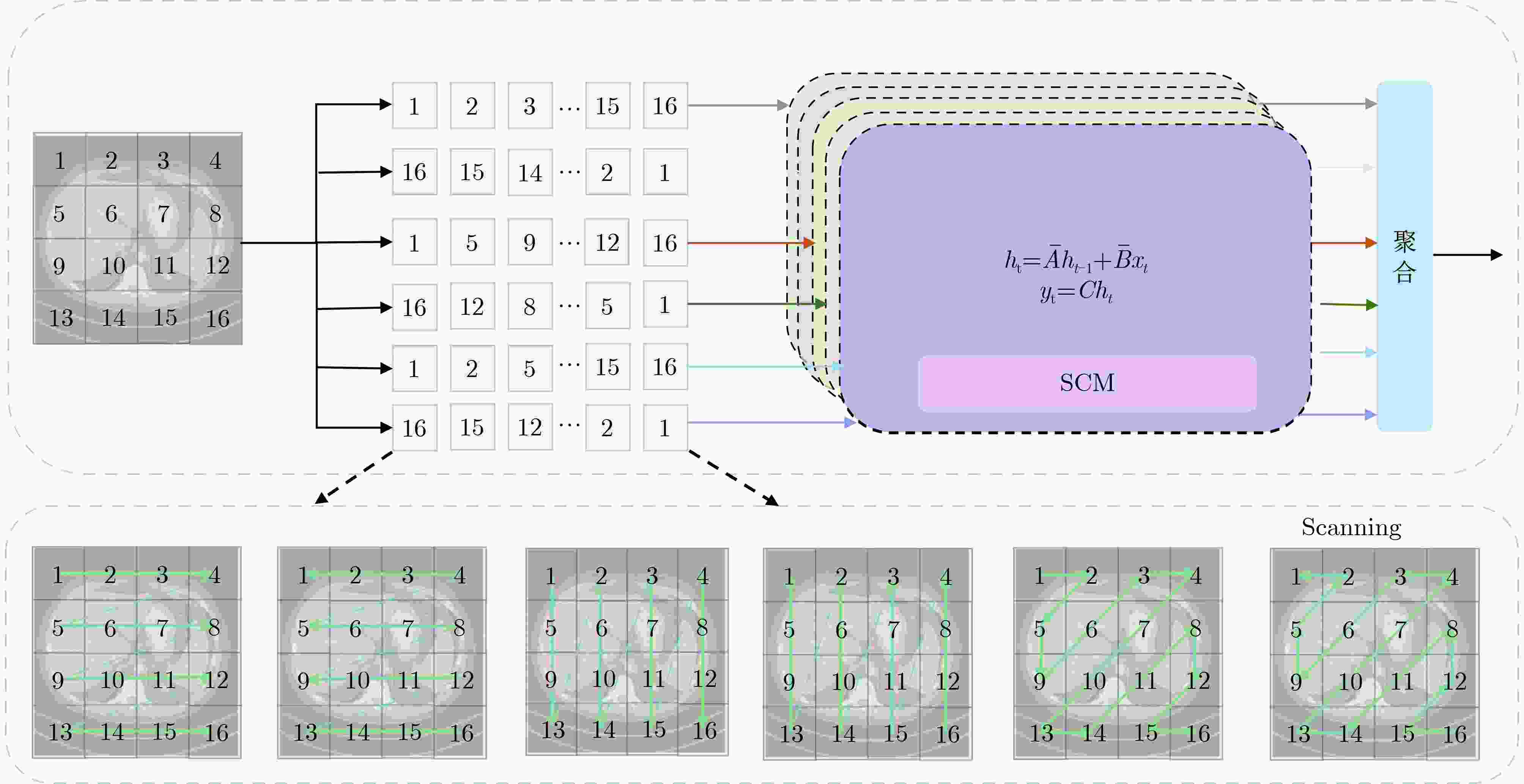
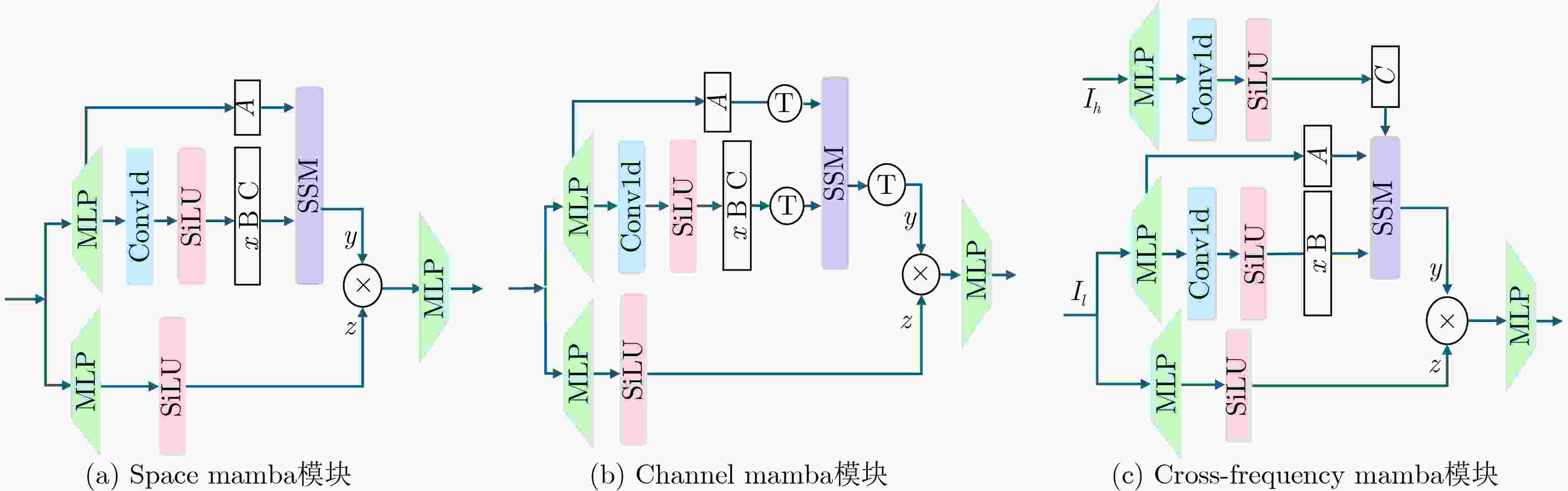
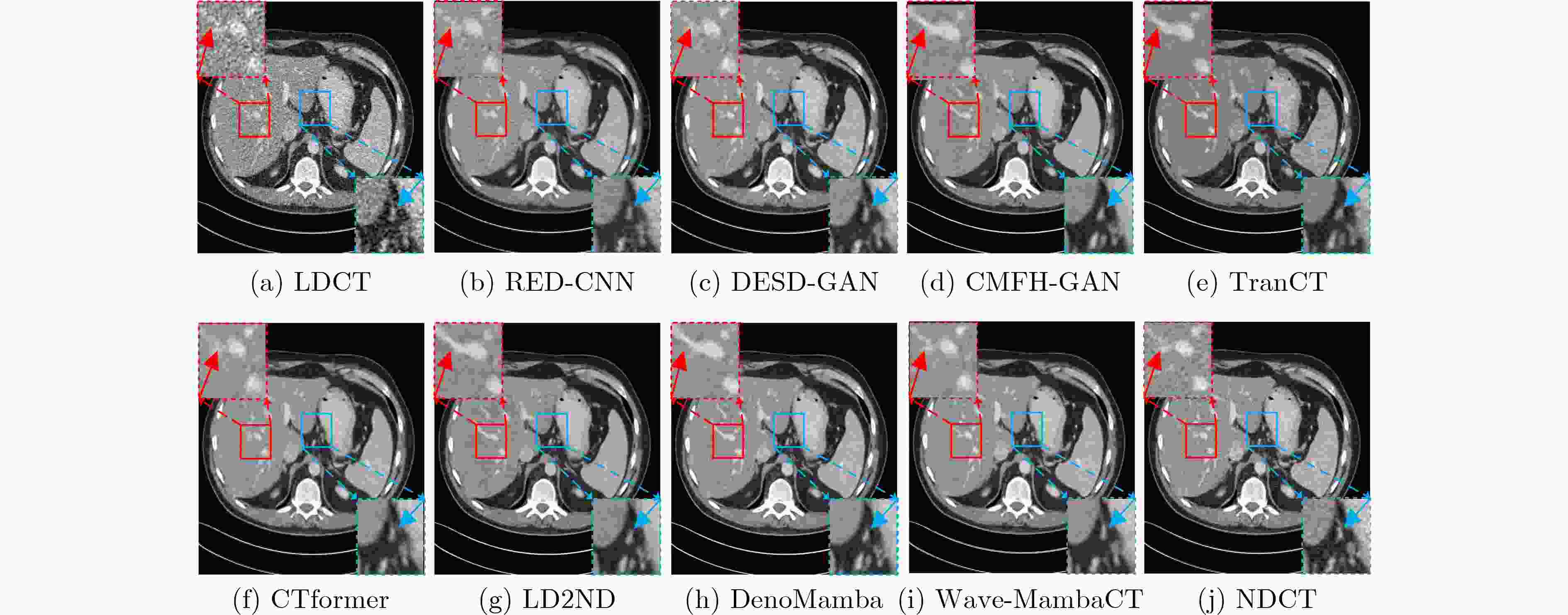
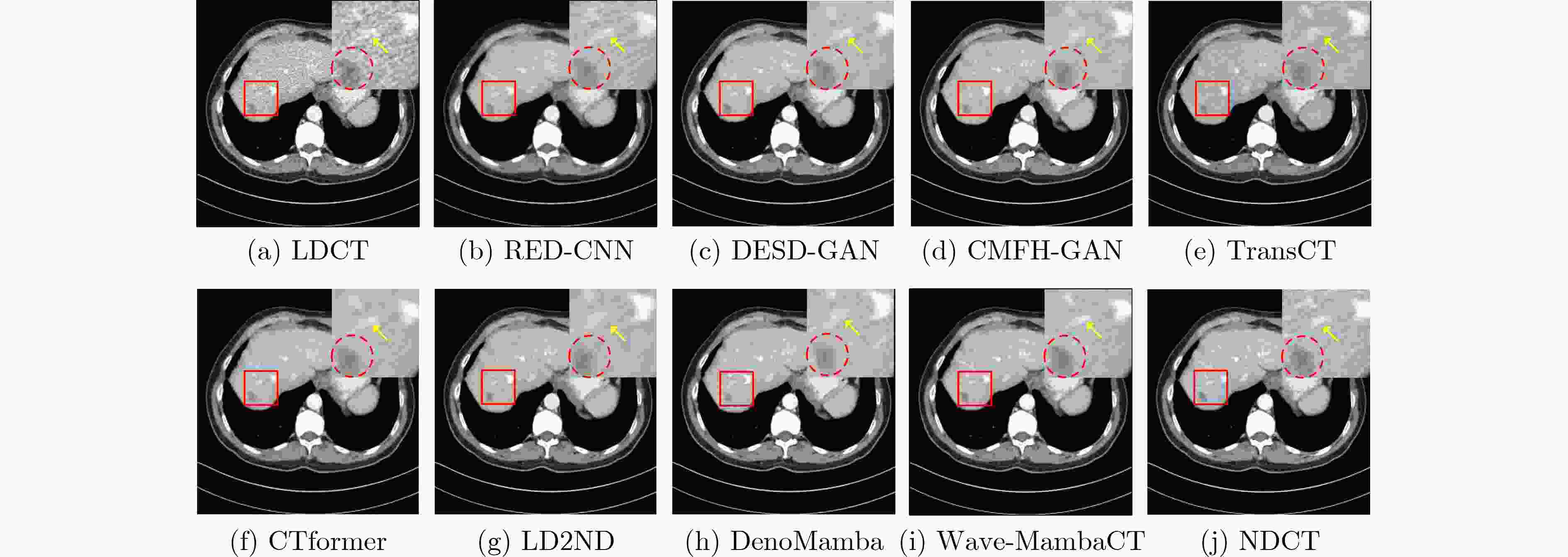
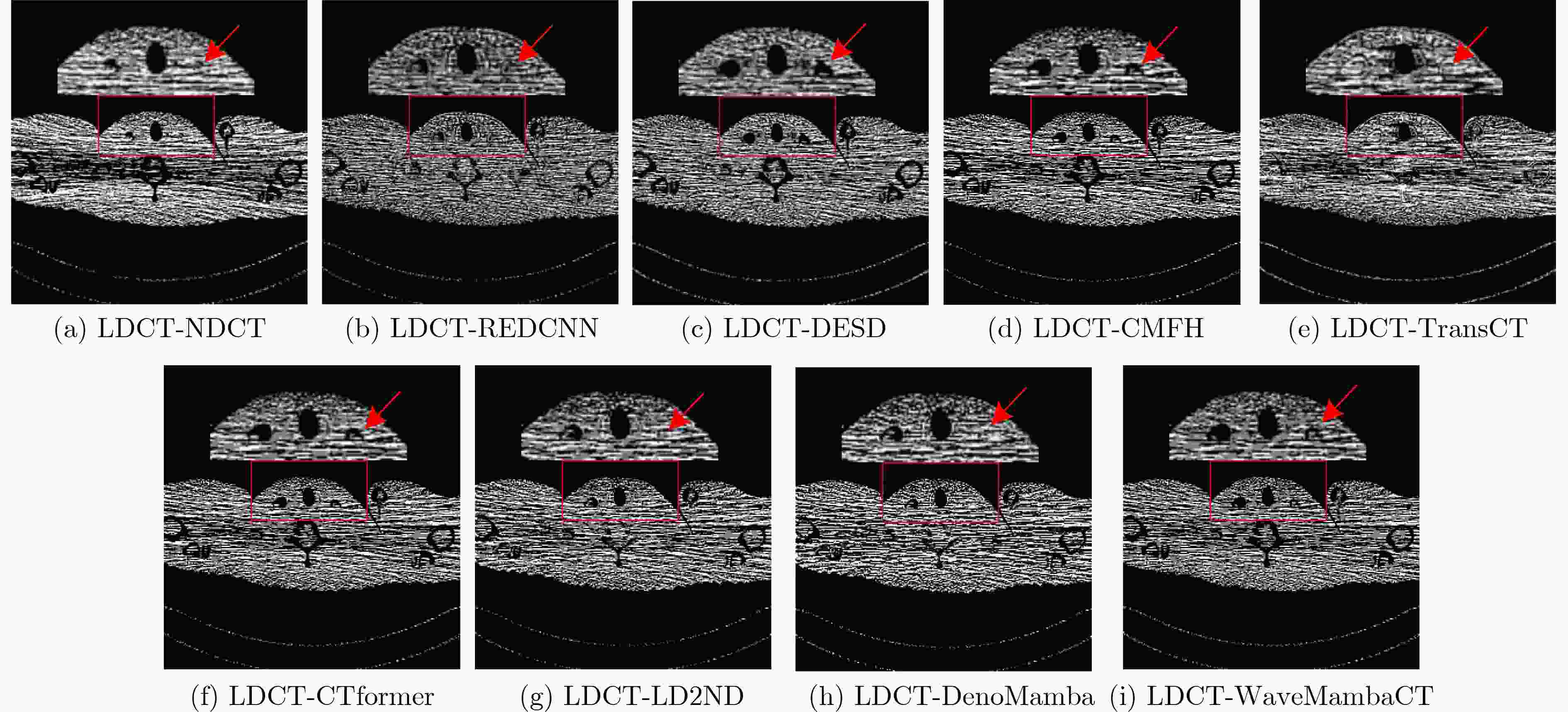
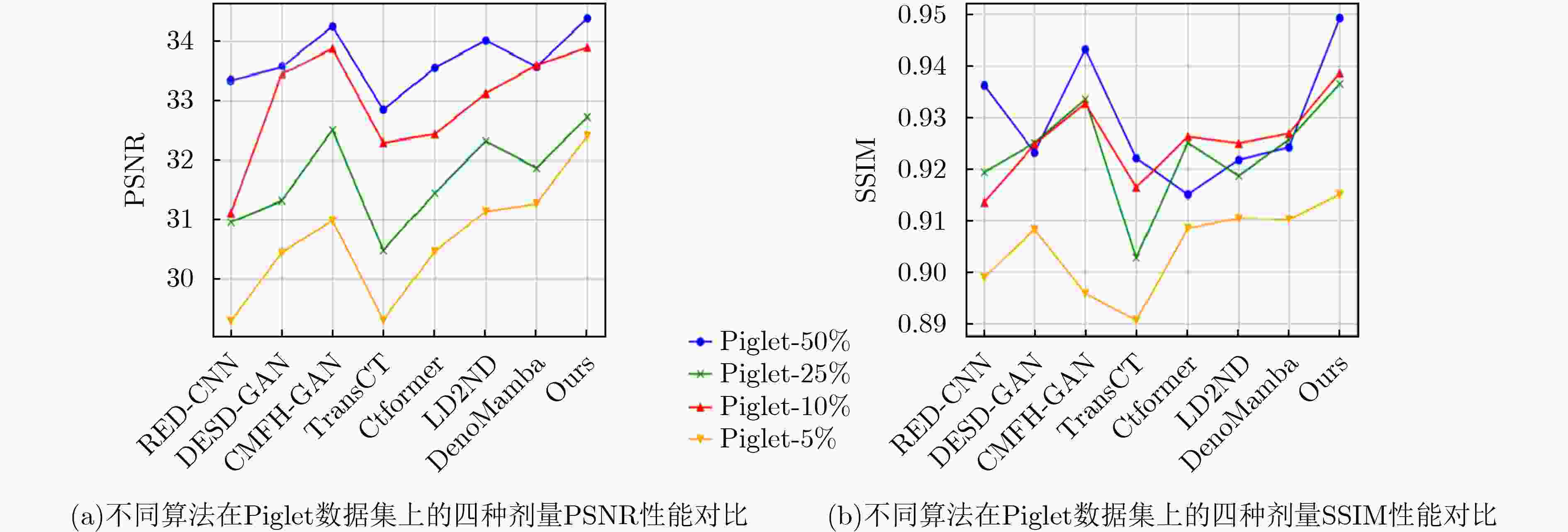
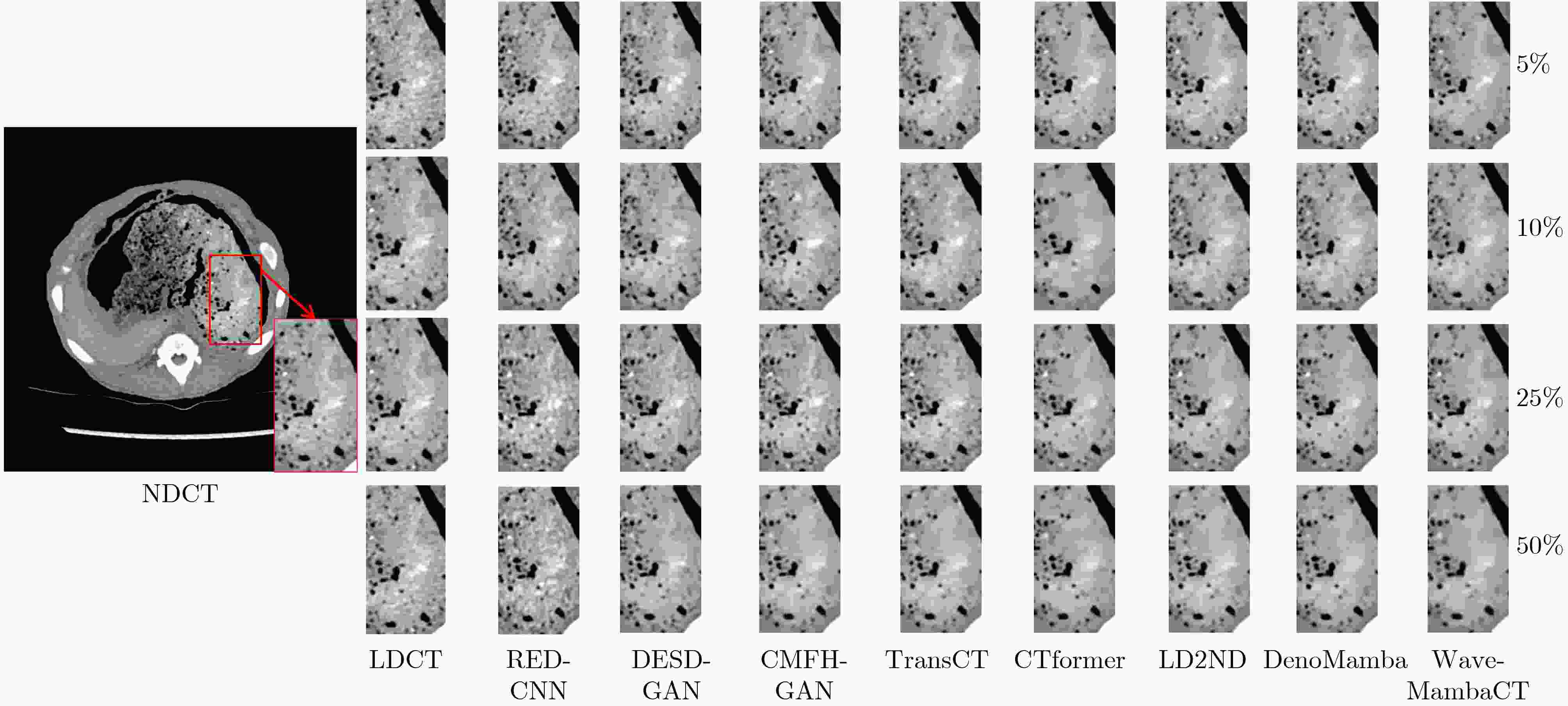
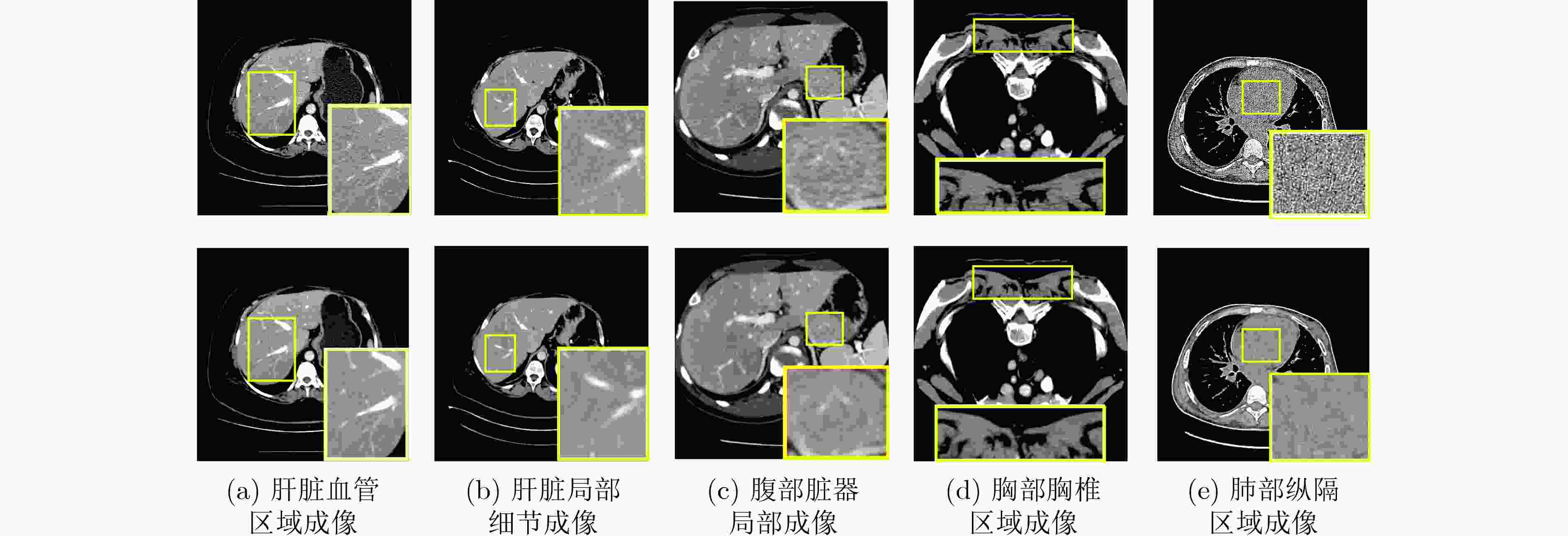
Objective Low-Dose Computed Tomography (LDCT) reduces patient radiation exposure but introduces substantial noise and artifacts into reconstructed images. Convolutional Neural Network (CNN)-based denoising approaches are limited by local receptive fields, which restrict their abilities to capture long-range dependencies. Transformer-based methods alleviate this limitation but incur quadratic computational complexity relative to image size. In contrast, State Space Model (SSM)–based Mamba frameworks achieve linear complexity for long-range interactions. However, existing Mamba-based methods often suffer from information loss and insufficient noise suppression. To address these limitations, we propose the Wave-MambaCT model. Methods The proposed Wave-MambaCT model adopts a multi-scale framework that integrates Discrete Wavelet Transform (DWT) with a Mamba module based on the SSM. First, DWT performs a two-level decomposition of the LDCT image, decoupling noise from Low-Frequency (LF) content. This design directs denoising primarily toward the High-Frequency (HF) components, facilitating noise suppression while preserving structural information. Second, a residual module combined with a Spatial-Channel Mamba (SCM) module extracts both local and global features from LF and HF bands at different scales. The noise-free LF features are then used to correct and enhance the corresponding HF features through an attention-based Cross-Frequency Mamba (CFM) module. Finally, inverse wavelet transform is applied in stages to progressively reconstruct the image. To further improve denoising performance and network stability, multiple loss functions are employed, including L1 loss, wavelet-domain LF loss, and adversarial loss for HF components. Results and Discussions Extensive experiments on the simulated Mayo Clinic datasets, the real Piglet datasets, and the hospital clinical dataset DeepLesion show that Wave-MambaCT provides superior denoising performance and generalization. On the Mayo dataset, a PSNR of 31.6528 is achieved, which is higher than that of the suboptimal method DenoMamba (PSNR31.4219 ), while MSE is reduced to0.00074 and SSIM and VIF are improved to0.8851 and0.4629 , respectively (Table 1 ). Visual results (Figs. 4 –6 ) demonstrate that edges and fine details such as abdominal textures and lesion contours are preserved, with minimal blurring or residual artifacts compared with competing methods. Computational efficiency analysis (Table 2 ) indicates that Wave-MambaCT maintains low FLOPs (17.2135 G) and parameters (5.3913 M). FLOPs are lower than those of all networks except RED-CNN, and the parameter count is higher only than those of RED-CNN and CTformer. During training, 4.12 minutes per epoch are required, longer only than RED-CNN. During testing,0.1463 seconds are required per image, which is at a medium level among the compared methods. Generalization tests on the Piglet datasets (Figs. 7 ,8 ,Tables 3 ,4 ) and DeepLesion (Fig. 9 ) further confirm the robustness and generalization capacity of Wave-MambaCT.In the proposed design, HF sub-bands are grouped, and noise-free LF information is used to correct and guide their recovery. This strategy is based on two considerations. First, it reduces network complexity and parameter count. Second, although the sub-bands correspond to HF information in different orientations, they are correlated and complementary as components of the same image. Joint processing enhances the representation of HF content, whereas processing them separately would require a multi-branch architecture, inevitably increasing complexity and parameters. Future work will explore approaches to reduce complexity and parameters when processing HF sub-bands individually, while strengthening their correlations to improve recovery. For structural simplicity, SCM is applied to both HF and LF feature extraction. However, redundancy exists when extracting LF features, and future studies will explore the use of different Mamba modules for HF and LF features to further optimize computational efficiency.Conclusions Wave-MambaCT integrates DWT for multi-scale decomposition, a residual module for local feature extraction, and an SCM module for efficient global dependency modeling to address the denoising challenges of LDCT images. By decoupling noise from LF content through DWT, the model enables targeted noise removal in the HF domain, facilitating effective noise suppression. The designed RSCM, composed of residual blocks and SCM modules, captures fine-grained textures and long-range interactions, enhancing the extraction of both local and global information. In parallel, the Cross-band Enhancement Module (CEM) employs noise-free LF features to refine HF components through attention-based CFM, ensuring structural consistency across scales. Ablation studies ( Table 5 ) confirm the essential contributions of both SCM and CEM modules to maintaining high performance. Importantly, the model’s staged denoising strategy achieves a favorable balance between noise reduction and structural preservation, yielding robustness to varying radiation doses and complex noise distributions.-
Key words:
- Low-dose CT /
- Artifact suppression /
- Wavelet transform /
- Mamba
-
表 1 不同降噪算法在Mayo测试集上的定量指标
方法 PSNR↑ SSIM↑ VIF↑ MSE↓ LDCT 26.7891 ±1.9782 0.8244 ±0.0503 0.3642 ±0.0580 0.00232 +0.00105 RED-CNN(2018) 31.0990 ±1.7724 0.8773 ±0.0390 0.4114 ±0.0568 0.00084 ±0.00036 DESD-GAN(2022) 30.8887 ±2.0041 0.8789 ±0.0406 0.3958 ±0.0514 0.00091 ±0.00049 CFMH-GAN(2023) 30.4322 ±2.5962 0.8703 ±0.0447 0.3521 ±0.0653 0.00098 ±0.00027 TransCT(2021) 30.5113 ±1.5217 0.8724 ±0.0386 0.3844 ±0.0513 0.00094 ±0.00033 CTformer(2023) 30.5176 ±2.8725 0.8764 ±0.0366 0.3895 ±0.0496 0.00095 ±0.00031 LD2ND(2024) 31.2681 ±1.5187 0.8811 ±0.0351 0.4185 ±0.0456 0.00081 ±0.00035 DenoMamba(2024) 31.4219 ±1.7312 0.8835 ±0.0391 0.4326 ±0.0672 0.00082 ±0.00032 Wave-MambaCT 31.6528 ±1.6959 0.8851 ±0.0391 0.4629 ±0.0547 0.00074 ±0.00031 表 2 不同降噪网络的指标对比
RED-CNN DESD-GAN CFMH-GAN TransCT CTformer LD2ND DenoMamba 本文 FLOPs(G) 5.0861 156.0901 30.1766 38.6857 18.1652 112.1565 31.1652 17.2135 Params(M) 0.4755 36.8598 46.3634 7.8608 1.4568 13.83 6.3403 5.3913 训练时间(min/epoch) 3.72 4.88 4.74 5.90 5.72 5.48 4.62 4.12 测试时间(s/张) 0.2190 0.1478 0.1291 0.1252 0.2287 0.1426 0.1513 0.1463 表 3 Piglet 作为训练集Mayo作为测试集时不同降噪算法的定量指标
方法 PSNR↑ SSIM↑ VIF↑ MSE↓ RED-CNN 30.0628 ±1.7213 0.8725 ±0.0411 0.4028 ±0.0568 0.00086 ±0.00032 DESD-GAN 30.3409 ±1.9024 0.8789 ±0.0406 0.3867 ±0.0547 0.00093 ±0.00041 CFMH-GAN 30.4258 ±2.2897 0.8684 ±0.0439 0.3509 ±0.0653 0.00097 ±0.00029 TransCT 30.4974 ±1.6866 0.8709 ±0.0423 0.3762 ±0.0556 0.00096 ±0.00031 CTformer 30.5032 ±2.3696 0.8727 ±0.0377 0.3848 ±0.0581 0.00098 ±0.00044 LD2ND 31.1396 ±1.5978 0.8795 ±0.0349 0.4123 ±0.0515 0.00087 ±0.00037 DenoMamba 31.2989 ±1.7121 0.8814 ±0.0326 0.4279 ±0.0537 0.00083 ±0.00036 Wave-MambaCT 31.5983 ±1.5216 0.8839 ±0.0315 0.4592 ±0.0547 0.00076 ±0.00033 表 4 Mayo作为训练集Piglet作为测试集时不同降噪算法的定量指标
方法 PSNR↑ SSIM↑ VIF↑ MSE↓ RED-CNN 31.4024 ±1.5122 0.9027 ±0.0175 0.4097 ±0.0468 0.00081 ±0.00038 DESD-GAN 32.1595 ±1.4358 0.9139 ±0.0211 0.4303 ±0.0511 0.00076 ±0.00034 CMFH-GAN 32.8126 ±1.3738 0.9233 ±0.0173 0.4369 ±0.0499 0.00069 ±0.00030 TransCT 31.1629 ±1.4127 0.9037 ±0.0128 0.4178 ±0.0512 0.00087 ±0.00035 CTformer 32.1805 ±1.2748 0.9146 ±0.0129 0.4417 ±0.0556 0.00077 ±0.00037 LD2ND 32.4385 ±1.3039 0.9194 ±0.0137 0.4432 ±0.0569 0.00070 ±0.00034 DenoMamba 33.0016 ±1.3076 0.9233 ±0.0128 0.4461 ±0.0582 0.00066 ±0.00029 Wave-MambaCT 33.2817 ±1.2037 0.9319 ±0.0131 0.4496 ±0.0529 0.00062 ±0.00031 表 5 在Mayo数据集上的消融实验
Method PSNR↑ SSIM↑ VIF↑ MSE↓ A 31.4121 ±1.6476 0.8817 ±0.0391 0.4459 ±0.0270 0.00079 ±0.00028 B 31.5284 ±1.6782 0.8845 ±0.0385 0.4456 ±0.0258 0.00077 ±0.00030 C 31.5741 ±1.7367 0.8849 ±0.0391 0.4619 ±0.0272 0.00075 ±0.00029 D 31.5431 ±1.7321 0.8846 ±0.0392 0.4598 ±0.0256 0.00076 ±0.00032 本文 31.6528 ±1.6959 0.8851 ±0.0391 0.4629 ±0.0547 0.00074 ±0.00029 表 6 在Mayo数据集上提出网络中模块数量的对比试验
方法 第1阶段 第2阶段 PSNR SSIM FLOPs(G) Params(M) 1 3 3 31.2542 ±1.7098 0.8761 ±0.0371 15.9218 4.7142 2(本文) 4 4 31.6528 ±1.6959 0.8851 ±0.0391 17.2135 5.3913 3 5 5 31.4813 ±1.6546 0.8750 ±0.0405 21.6529 5.8974 表 7 损失函数中不同系数比率的定量结果
系数比例 PSNR SSIM 系数比例 PSNR SSIM 1:0.05:0.10 31.2517 0.8721 1:0.01:0.15 31.4813 0.8812 1:0.05:0.15 31.6528 0.8851 1:0.05:0.15 31.6528 0.8851 1:0.05:0.20 31.3219 0.8782 1:0.10:0.15 31.5235 0.8795 -
[1] 张权. 低剂量X线CT重建若干问题研究[D]. [博士论文], 东南大学, 2015.ZHANG Quan. A study on some problems in image reconstruction for low-dose CT system[D]. [Ph. D. dissertation], Southeast University, 2015. [2] DE BASEA M B, THIERRY-CHEF I, HARBRON R, et al. Risk of hematological malignancies from CT radiation exposure in children, adolescents and young adults[J]. Nature Medicine, 2023, 29(12): 3111–3119. doi: 10.1038/s41591-023-02620-0. [3] CHEN Hu, ZHANG Yi, KALRA M K, et al. Low-dose CT with a Residual Encoder-Decoder Convolutional Neural Network (RED-CNN)[J]. IEEE Transactions on Medical Imaging, 2017, 36(12): 2524–2535. doi: 10.1109/TMI.2017.2715284. [4] LIANG Tengfei, JIN Yi, LI Yidong, et al. EDCNN: Edge enhancement-based densely connected network with compound loss for low-dose CT denoising[C]. The 15th IEEE International Conference on Signal Processing, Beijing, China, 2020: 193–198, doi: 10.1109/ICSP48669.2020.9320928. [5] SAIDULU N and MUDULI P R. Asymmetric convolution-based GAN framework for low-dose CT image denoising[J]. Computers in Biology and Medicine, 2025, 190: 109965. doi: 10.1016/j.compbiomed.2025.109965. [6] 张雄, 杨琳琳, 上官宏, 等. 基于生成对抗网络和噪声水平估计的低剂量CT图像降噪方法[J]. 电子与信息学报, 2021, 43(8): 2404–2413. doi: 10.11999/JEIT200591.ZHANG Xiong, YANG Linlin, SHANGGUAN Hong, et al. A low-dose ct image denoising method based on generative adversarial network and noise level estimation[J]. Journal of Electronics & Information Technology, 2021, 43(8): 2404–2413. doi: 10.11999/JEIT200591. [7] HAN Zefang, SHANGGUAN Hong, ZHANG Xiong, et al. A dual-encoder-single-decoder based low-dose CT denoising network[J]. IEEE Journal of Biomedical and Health Informatics, 2022, 26(7): 3251–3260. doi: 10.1109/JBHI.2022.3155788. [8] HAN Zefang, SHANGGUAN Hong, ZHANG Xiong, et al. A coarse-to-fine multi-scale feature hybrid low-dose CT denoising network[J]. Signal Processing: Image Communication, 2023, 118: 117009. doi: 10.1016/j.image.2023.117009. [9] ZHAO Haoyu, GU Yuliang, ZHAO Zhou, et al. WIA-LD2ND: Wavelet-based image alignment for self-supervised low-dose CT denoising[C]. The 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 764–774. doi: 10.1007/978-3-031-72104-5_73. [10] LUTHRA A, SULAKHE H, MITTAL T, et al. Eformer: Edge enhancement based transformer for medical image denoising[J]. arXiv preprint arXiv: 2109.08044, 2021. [11] ZHANG Zhicheng, YU Lequan, LIANG Xiaokun, et al. TransCT: Dual-path transformer for low dose computed tomography[C]. The 24th International Conference on Medical Image Computing and Computer Assisted Intervention, Strasbourg, France, 2021: 55–64. doi: 10.1007/978-3-030-87231-1_6. [12] WANG Dayang, FAN Fenglei, WU Zhan, et al. CTformer: Convolution-free Token2Token dilated vision transformer for low-dose CT denoising[J]. Physics in Medicine & Biology, 2023, 68(6): 065012. doi: 10.1088/1361-6560/acc000. [13] JIAN Muwei, YU Xiaoyang, ZHANG Haoran, et al. SwinCT: Feature enhancement based low-dose CT images denoising with swin transformer[J]. Multimedia Systems, 2024, 30(1): 1. doi: 10.1007/s00530-023-01202-x. [14] LI Haoran, YANG Xiaomin, YANG Sihan, et al. Transformer with double enhancement for low-dose CT denoising[J]. IEEE Journal of Biomedical and Health Informatics, 2023, 27(10): 4660–4671. doi: 10.1109/JBHI.2022.3216887. [15] GU A and DAO T. Mamba: Linear-time sequence modeling with selective state spaces[J]. arXiv preprint arXiv: 2312.00752, 2024. [16] DAO T and GU A. Transformers are SSMs: Generalized models and efficient algorithms through structured state space duality[C]. The 41st International Conference on Machine Learning, Vienna, Austria, 2024: 399. [17] LIU Yue, TIAN Yunjie, ZHAO Yuzhong, et al. VMamba: Visual state space model[C]. The 38th International Conference on Neural Information Processing Systems, Vancouver, Canada, 2024: 3273. [18] öZTÜRK Ş, DURAN O C, and çUKUR T. DenoMamba: A fused state-space model for low-dose CT denoising[J]. arXiv Preprint arXiv: 2409.13094, 2024. [19] LI Linxuan, WEI Wenjia, YANG Luyao, et al. CT-Mamba: A hybrid convolutional state space model for low-dose CT denoising[J]. Computerized Medical Imaging and Graphics, 2025, 124: 102595. doi: 10.1016/j.compmedimag.2025.102595. [20] XU Guoping, LIAO Wentao, ZHANG Xuan, et al. Haar wavelet downsampling: A simple but effective downsampling module for semantic segmentation[J]. Pattern Recognition, 2023, 143: 109819. doi: 10.1016/j.patcog.2023.109819. [21] AAPM. Low dose CT grand challenge[EB/OL]. http://www.aapm.org/GrandChallenge/LowDoseCT/, 2017. [22] Piglet dataset[EB/OL]. https://universe.roboflow.com/piglet-dataset, 2025. [23] YAN Ke, WANG Xiaosong, LU Le, et al. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning[J]. Journal of Medical Imaging, 2018, 5(3): 036501. doi: 10.1117/1.JMI.5.3.036501. -





 下载:
下载:


 下载:
下载:
