Multi-Resolution Spatio-Temporal Fusion Graph Convolutional Network for Attention Deficit Hyperactivity Disorder Classification
-
摘要: 神经发育障碍疾病患者的精准分类是医学领域的一项重要挑战,对于疾病诊断和指导治疗至关重要。然而,现有基于图卷积网络(GCNs)的方法通常采用单一分辨率空间特征,忽视了多分辨率下的空间信息以及时间信息。为了克服上述局限性,该文提出一种多分辨率时空融合图卷积网络(MSTF-GCN)。在多个分辨率空间下构建多个大脑功能连通性网络,使用支持向量机-递归特征消除提取最优空间特征。为了保留全局时间信息并使网络具有捕获信号不同层次变化的能力,将全局时间信号及其差分信号输入到时间卷积网络中学习复杂时间维度的依赖关系,提取时间特征。结合时空信息构建群体图,利用多通道图卷积网络灵活地融合不同分辨率的群体图数据,最后融入非成像数据信息生成有效的多通道多类型时空融合分类特征,有效提升了MSTF-GCN模型的分类性能。将MSTF-GCN应用于注意力缺陷多动障碍(ADHD)患者分类识别,在ADHD-200数据集两个成像站点上的分类准确率分别达到了75.92%和82.95%,实验结果优于已有的流行算法,验证了MSTF-GCN的有效性。
-
关键词:
- 多分辨率时空融合图卷积网络 /
- 时空融合 /
- 多分辨率 /
- 注意力缺陷多动障碍
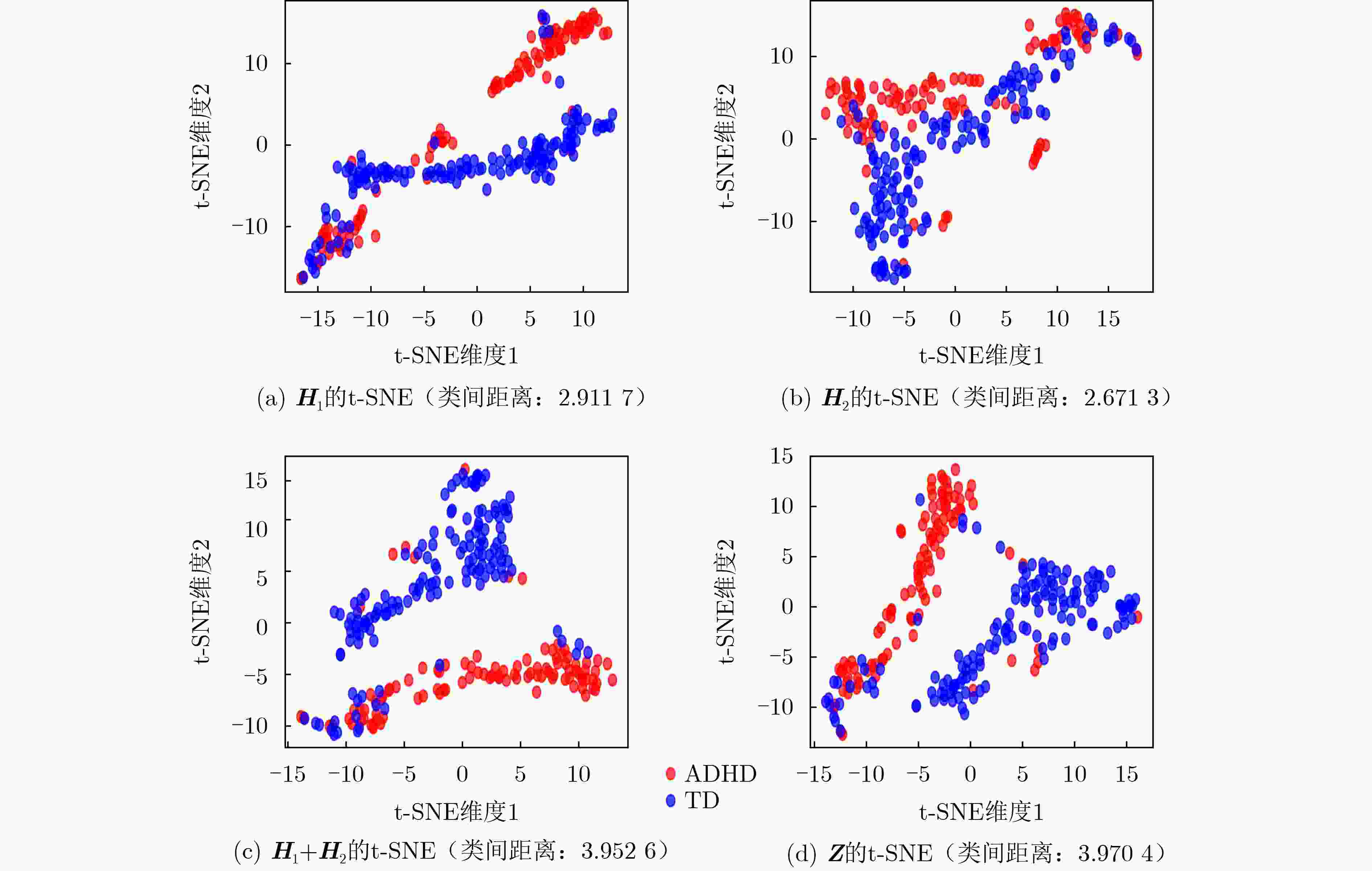
Abstract:Objective Predicting neurodevelopmental disorders remains a central challenge in neuroscience and artificial intelligence. Attention Deficit Hyperactivity Disorder (ADHD), a representative complex brain disorder, presents diagnostic difficulties due to its increasing prevalence, clinical heterogeneity, and reliance on subjective criteria, which impede early and accurate detection. Developing objective, data-driven classification models is therefore of significant clinical relevance. Existing graph convolutional network-based approaches for functional brain network analysis are constrained by several limitations. Most adopt single-resolution brain parcellation schemes, reducing their capacity to capture complementary features from multi-resolution functional Magnetic Resonance Imaging (fMRI) data. Moreover, the lack of effective cross-scale feature fusion restricts the integration of essential features across resolutions, hampering the modeling of hierarchical dependencies among brain regions. To address these limitations, this study proposes a Multi-resolution Spatio-Temporal Fusion Graph Convolutional Network (MSTF-GCN), which integrates spatiotemporal features across multiple fMRI resolutions. The proposed method substantially improves the accuracy and robustness of functional brain network classification for ADHD. Methods The MSTF-GCN improves learning performance through two main components: (1) construction of multi-resolution, multi-channel networks, and (2) comprehensive fusion of temporal and spatial information. Multiple brain atlases at different resolutions are employed to parcellate the brain and generate functional connectivity networks. Spatial features are extracted from these networks, and optimal nodal features are selected using Support Vector Machine-Recursive Feature Elimination (SVM-RFE). To preserve global temporal characteristics and capture hierarchical signal variations, both the original time series and their differential signals are processed using a temporal convolutional network. This structure enables the extraction of complex temporal features and inter-subject temporal correlations. Spatial features from different resolutions are then fused with temporal correlations to form population graphs, which are adaptively integrated via a multi-channel graph convolutional network. Non-imaging data are also integrated to produce effective multi-channel, multi-modal spatiotemporal fusion features. The final classification is performed using a fully connected layer. Results and Discussions The proposed MSTF-GCN model is evaluated for ADHD classification using two independent sites from the ADHD-200 dataset: Peking and NI. The model consistently outperforms existing methods, achieving classification accuracies of 75.92% at the Peking site and 82.95% at the NI site ( Table 2 ,Table 3 ). Ablation studies confirm the contributions of two key components: (1) The multi-atlas, multi-resolution feature extraction strategy significantly enhances classification accuracy (Table 4 ), supporting the utility of complementary cross-scale topological information; (2) The multimodal fusion strategy, which incorporates non-imaging variables (gender and age), yields notable performance improvements (Table 5 ). Furthermore, t-SNE visualization and inter-class distance analysis (Fig. 6 ) show that MSTF-GCN generates a feature space with clearer class separation, reflecting the effectiveness of its multi-channel spatiotemporal fusion design. Overall, the MSTF-GCN model achieves superior performance compared with state-of-the-art methods and demonstrates strong robustness across sites, offering a promising tool for auxiliary diagnosis of brain disorders.Conclusions This study proposes a novel multi-channel graph embedding framework that integrates spatial topological and temporal features derived from multi-resolution fMRI data, leading to marked improvements in classification performance. Experimental results show that the MSTF-GCN method exceeds current state-of-the-art algorithms, with accuracy gains of 3.92% and 8.98% on the Peking and NI sites, respectively. These findings confirm its strong performance and cross-site robustness in ADHD classification. Future work will focus on constructing more expressive hypergraph neural networks to capture higher-order relationships within functional brain networks. -
表 1 实验中使用的受试者数据相关信息
站点 样本(ADHD/TD) 时间序列 性别(女/男) 年龄 均值$ \pm $标准差 最大/最小 Peking 245(102/143) 235 71/174 11.216$ \pm $1.973 17/8 NI 72(35/37) 260 30/42 17.222$ \pm $3.073 26/11 表 2 本文方法与已有流行方法在ADHD-200数据集Peking站点上的二分类实验结果比较(%)
表 3 本文方法与已有流行方法在ADHD-200数据集NI站点上的二分类实验结果比较(%)
表 4 多分辨率对实验结果的影响(%)
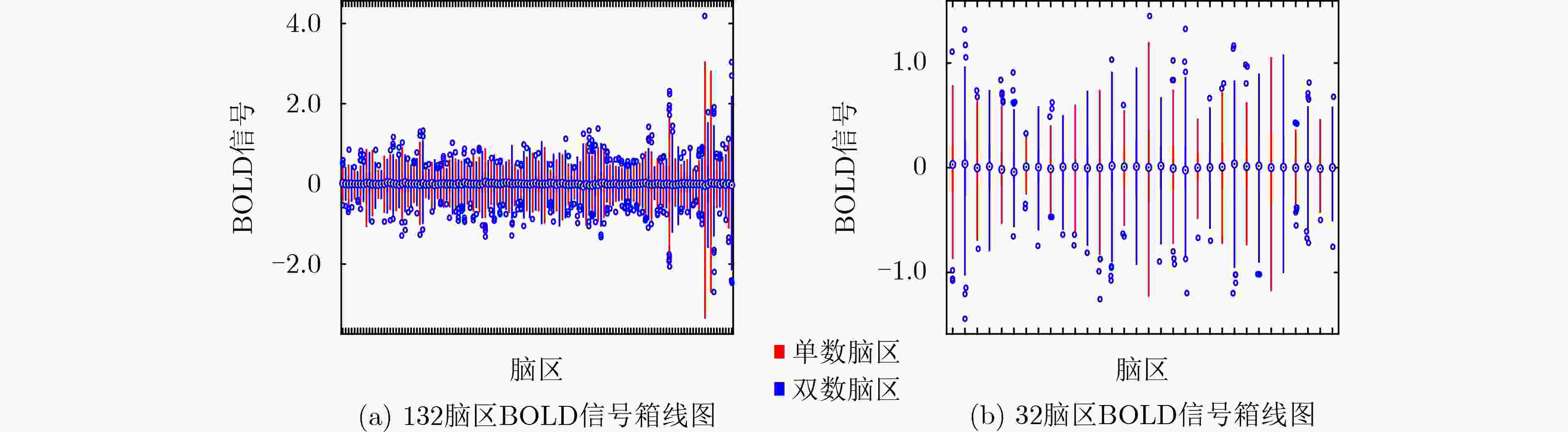
脑图谱 Acc(Var) Auc(Var) F1(Var) N1 = 132 73.49(0.25) 72.34(0.92) 77.17(0.25) N2 = 32 70.20(0.13) 67.92(0.43) 76.93(0.05) N1 + N2 75.92(0.06) 74.22(0.42) 80.49(0.04) 表 5 非成像数据对实验结果的影响(%)
非成像数据 Acc(Var) Auc(Var) F1(Var) 无 73.06(0.26) 71.49(0.49) 78.50(0.11) 性别 73.47(0.13) 72.56(0.38) 78.39(0.14) 年龄 74.29(0.08) 72.67(0.45) 79.60(0.09) 性别和年龄 75.92(0.06) 74.22(0.42) 80.49(0.04) -
[1] SHARMA A and COUTURE J. A review of the pathophysiology, etiology, and treatment of attention-deficit hyperactivity disorder (ADHD)[J]. Annals of Pharmacotherapy, 2014, 48(2): 209–225. doi: 10.1177/1060028013510699. [2] 杨健, 苗硕. 注意缺陷多动障碍患儿认知功能检测方法的进展[J]. 北京医学, 2015, 37(6): 507–508. doi: 10.15932/j.0253-9713.2015.6.001.YANG Jian and MIAO Shuo. Advances in detection methods of cognitive function in children with attention deficit hyperactivity disorder[J]. Beijing Medical Journal, 2015, 37(6): 507–508. doi: 10.15932/j.0253-9713.2015.6.001. [3] American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders[M]. 5th ed. Washington: American Psychiatric Publishing, 2013: 59–65. doi: 10.1176/appi.books.9780890425596. [4] ARBABSHIRANI M R, PLIS S, SUI Jing, et al. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls[J]. NeuroImage, 2017, 145: 137–165. doi: 10.1016/j.neuroimage.2016.02.079. [5] GRIMM O, VAN ROOIJ D, HOOGMAN M, et al. Transdiagnostic neuroimaging of reward system phenotypes in ADHD and comorbid disorders[J]. Neuroscience & Biobehavioral Reviews, 2021, 128: 165–181. doi: 10.1016/J.NEUBIOREV.2021.06.025. [6] JIE Biao, LIU Mingxia, and SHEN Dinggang. Integration of temporal and spatial properties of dynamic connectivity networks for automatic diagnosis of brain disease[J]. Medical Image Analysis, 2018, 47: 81–94. doi: 10.1016/j.media.2018.03.013. [7] 杨昆, 常世龙, 王尉丞, 等. 基于sECANet通道注意力机制的肾透明细胞癌病理图像ISUP分级预测[J]. 电子与信息学报, 2022, 44(1): 138–148. doi: 10.11999/JEIT210900.YANG Kun, CHANG Shilong, WANG Yucheng, et al. Predict the ISUP grade of clear cell renal cell carcinoma using pathological images based on sECANet chanel attention[J]. Journal of Electronics & Information Technology, 2022, 44(1): 138–148. doi: 10.11999/JEIT210900. [8] 金怀平, 薛飞跃, 李振辉, 等. 基于病理图像集成深度学习的胃癌预后预测方法[J]. 电子与信息学报, 2023, 45(7): 2623–2633. doi: 10.11999/JEIT220655.JIN Huaiping, XUE Feiyue, LI Zhenhui, et al. Prognostic prediction of gastric cancer based on ensemble deep learning of pathological images[J]. Journal of Electronics & Information Technology, 2023, 45(7): 2623–2633. doi: 10.11999/JEIT220655. [9] PARISOT S, KTENA S I, FERRANTE E, et al. Spectral graph convolutions for population-based disease prediction[C]. The 20th International Conference on Medical Image Computing and Computer Assisted Intervention, Quebec City, Canada, 2017: 177–185. doi: 10.1007/978-3-319-66179-7_21. [10] KAZI A, SHEKARFOROUSH S, ARVIND KRISHNA S, et al. InceptionGCN: Receptive field aware graph convolutional network for disease prediction[C]. The 26th International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2019: 73–85. doi: 10.1007/978-3-030-20351-1_6. [11] JIANG Hao, CAO Peng, XU MingYi, et al. Hi-GCN: A hierarchical graph convolution network for graph embedding learning of brain network and brain disorders prediction[J]. Computers in Biology and Medicine, 2020, 127: 104096. doi: 10.1016/j.compbiomed.2020.104096. [12] LI Lanting, JIANG Hao, WEN Guangqi, et al. TE-HI-GCN: An ensemble of transfer hierarchical graph convolutional networks for disorder diagnosis[J]. Neuroinformatics, 2022, 20(2): 353–375. doi: 10.1007/S12021-021-09548-1. [13] HUANG Yongxiang and CHUNG A C S. Disease prediction with edge-variational graph convolutional networks[J]. Medical Image Analysis, 2022, 77: 102375. doi: 10.1016/J.MEDIA.2022.102375. [14] PARK K W and CHO S B. A residual graph convolutional network with spatio-temporal features for autism classification from fMRI brain images[J]. Applied Soft Computing, 2023, 142: 110363. doi: 10.1016/j.asoc.2023.110363. [15] LI Ziyu, LI Qing, ZHU Zhiyuan, et al. Multi-scale spatio-temporal fusion with adaptive brain topology learning for fMRI based neural decoding[J]. IEEE Journal of Biomedical and Health Informatics, 2024, 28(1): 262–272. doi: 10.1109/JBHI.2023.3327023. [16] LIU Rui, HUANG Zhian, HU Yao, et al. Spatio-temporal hybrid attentive graph network for diagnosis of mental disorders on fMRI time-series data[J]. IEEE Transactions on Emerging Topics in Computational Intelligence, 2024, 8(6): 4046–4058. doi: 10.1109/TETCI.2024.3386612. [17] MITRA A, SNYDER A Z, HACKER C D, et al. Lag structure in resting-state fMRI[J]. Journal of Neurophysiology, 2014, 111(11): 2374–2391. doi: 10.1152/jn.00804.2013. [18] WU Zonghan, PAN Shirui, LONG Guodong, et al. Graph WaveNet for deep spatial-temporal graph modeling[C]. The Twenty-Eighth International Joint Conference on Artificial Intelligence, Macao, China, 2019: 1907–1913. doi: 10.24963/ijcai.2019/264. [19] ASHBURNER J. SPM: A history[J]. NeuroImage, 2012, 62(2): 791–800. doi: 10.1016/j.neuroimage.2011.10.025. [20] WHITFIELD-GABRIELI S and NIETO-CASTANON A. CONN: A functional connectivity toolbox for correlated and anticorrelated brain networks[J]. Brain Connectivity, 2012, 2(3): 125–141. doi: 10.1089/brain.2012.0073. [21] KINGMA D P and BA J. Adam: A method for stochastic optimization[C]. The 3rd International Conference on Learning Representations, San Diego, USA, 2015. [22] RIAZ A, ASAD M, ALONSO E, et al. DeepFMRI: End-to-end deep learning for functional connectivity and classification of ADHD using fMRI[J]. Journal of Neuroscience Methods, 2020, 335: 108506. doi: 10.1016/j.jneumeth.2019.108506. [23] DOU Chengfeng, ZHANG Shikun, WANG Hanping, et al. ADHD fMRI short-time analysis method for edge computing based on multi-instance learning[J]. Journal of Systems Architecture, 2020, 111: 101834. doi: 10.1016/j.sysarc.2020.101834. [24] ZHAO Kanhao, DUKA B, XIE Hua, et al. A dynamic graph convolutional neural network framework reveals new insights into connectome dysfunctions in ADHD[J]. NeuroImage, 2022, 246: 118774. doi: 10.1016/j.neuroimage.2021.118774. [25] KIM B, PARK J, KIM T, et al. Finding essential parts of the brain in rs-fMRI can improve ADHD diagnosis using deep learning[J]. IEEE Access, 2023, 11: 116065–116075. doi: 10.1109/ACCESS.2023.3324670. [26] PEI Shengbing, HE Fan, CAO Shuai, et al. Learning meta-stable state transition representation of brain function for ADHD identification[J]. IEEE Transactions on Instrumentation and Measurement, 2023, 72: 2530713. doi: 10.1109/TIM.2023.3324338. -





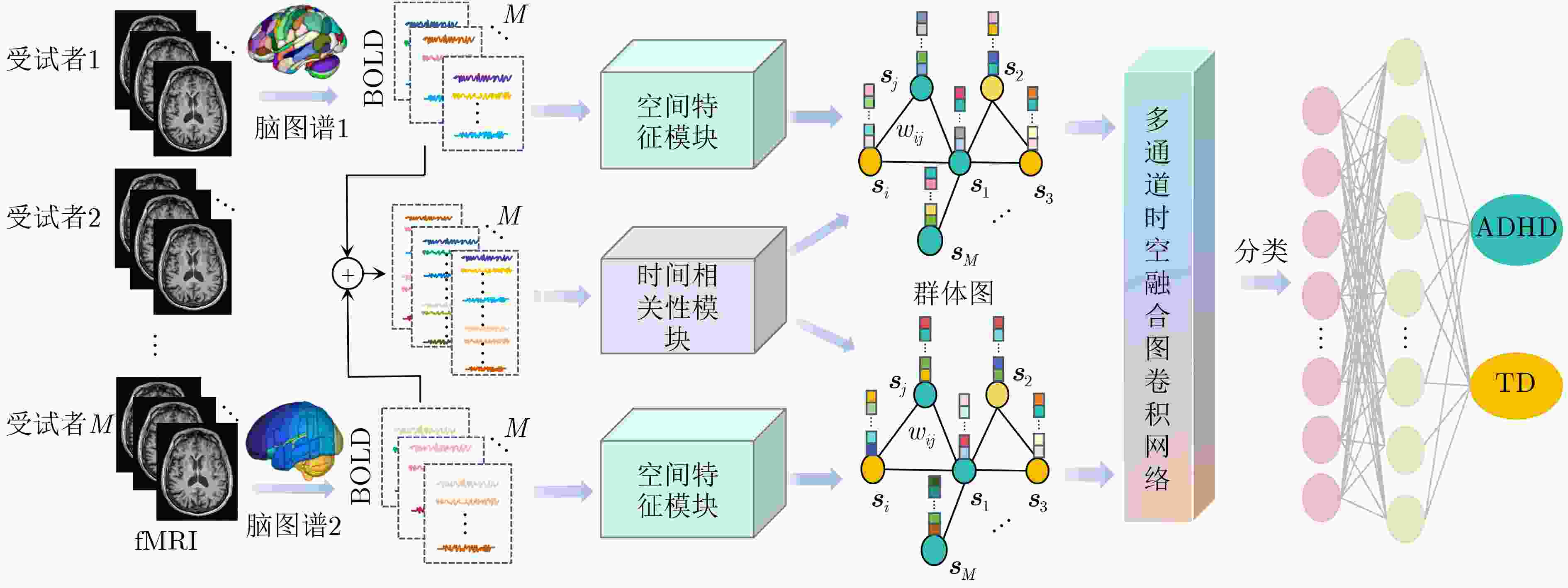
 下载:
下载:
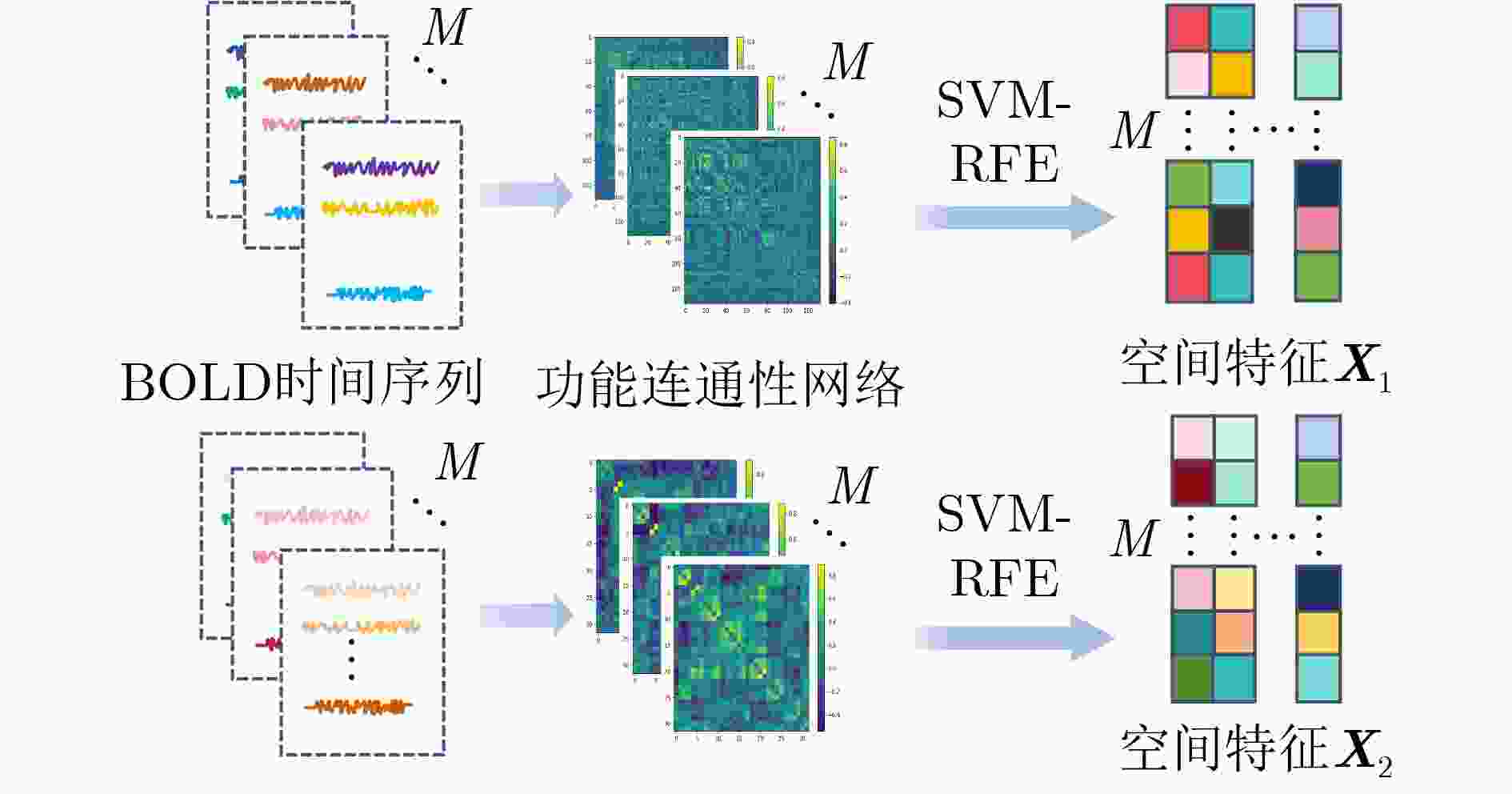
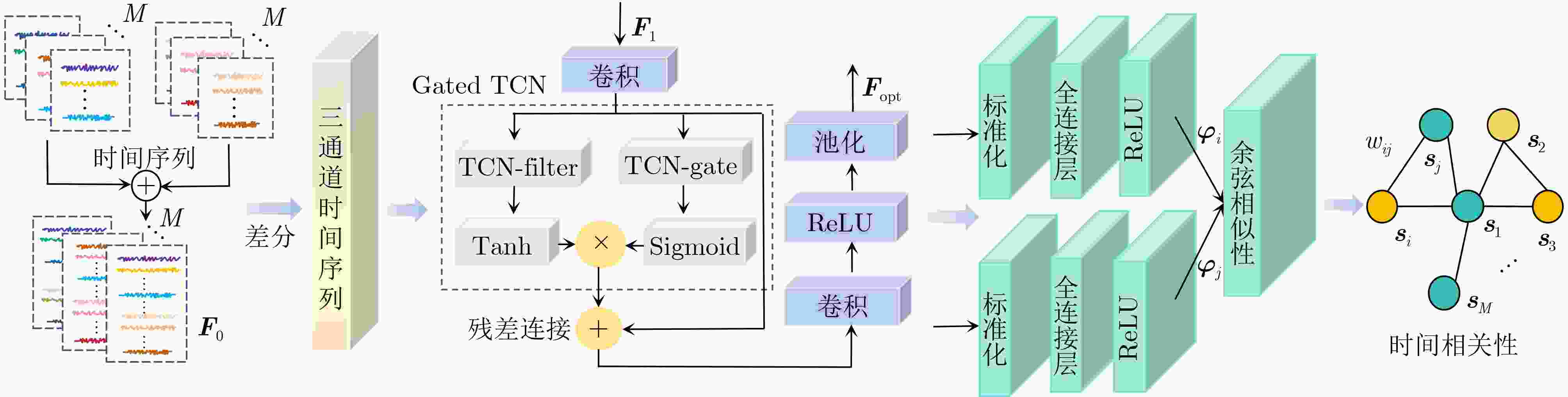
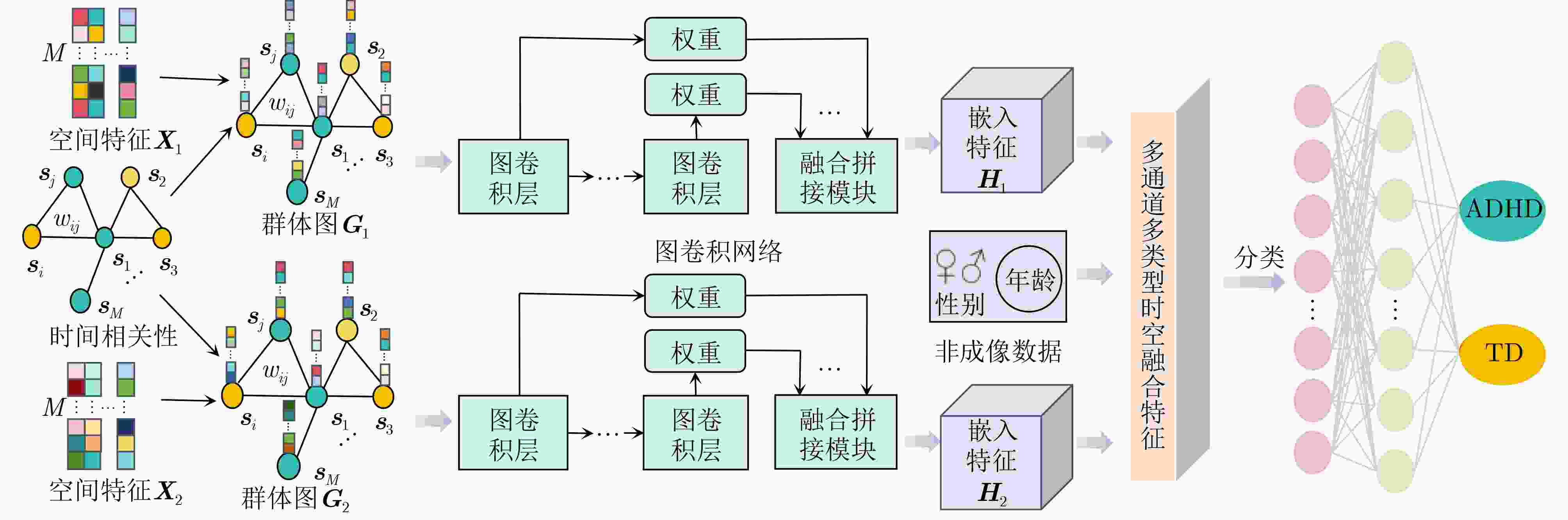


 下载:
下载:
