A Medical Video Segmentation Algorithm Integrating Neighborhood Attention and State Space Model
-
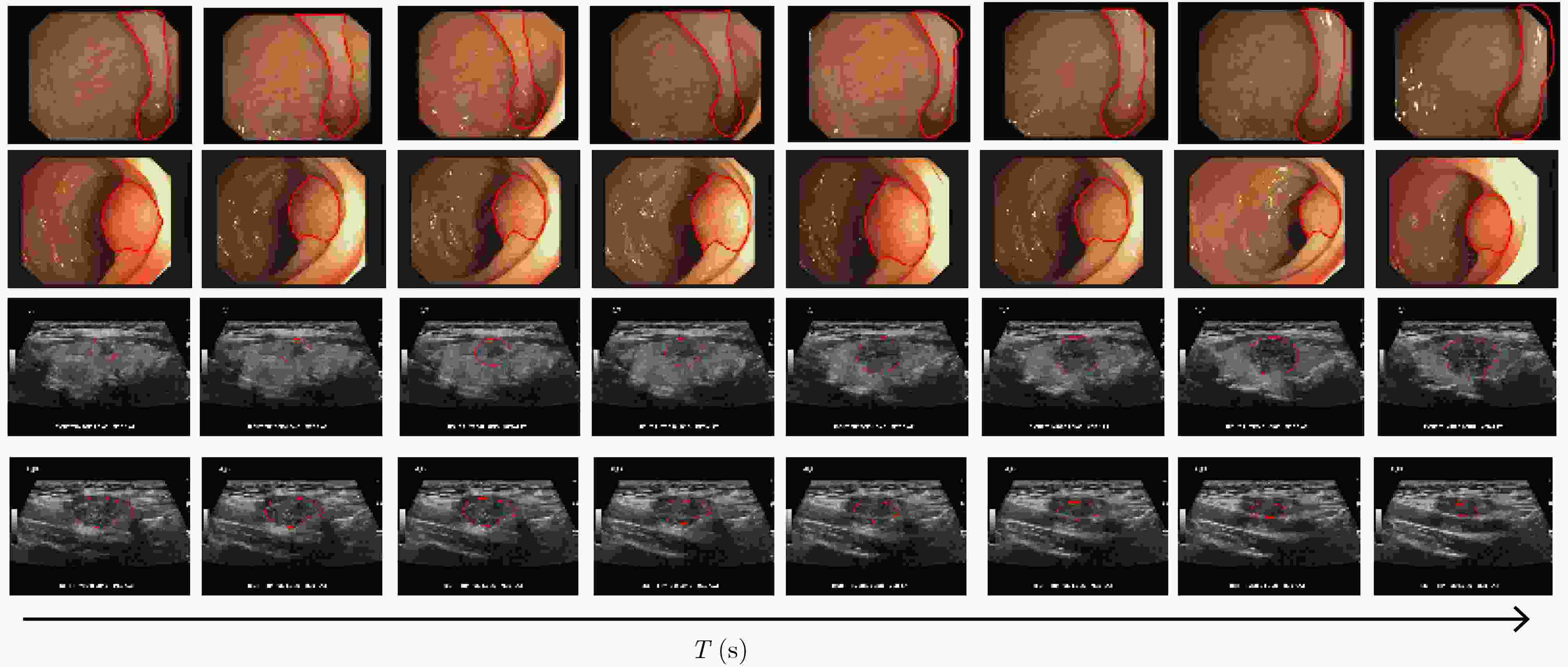
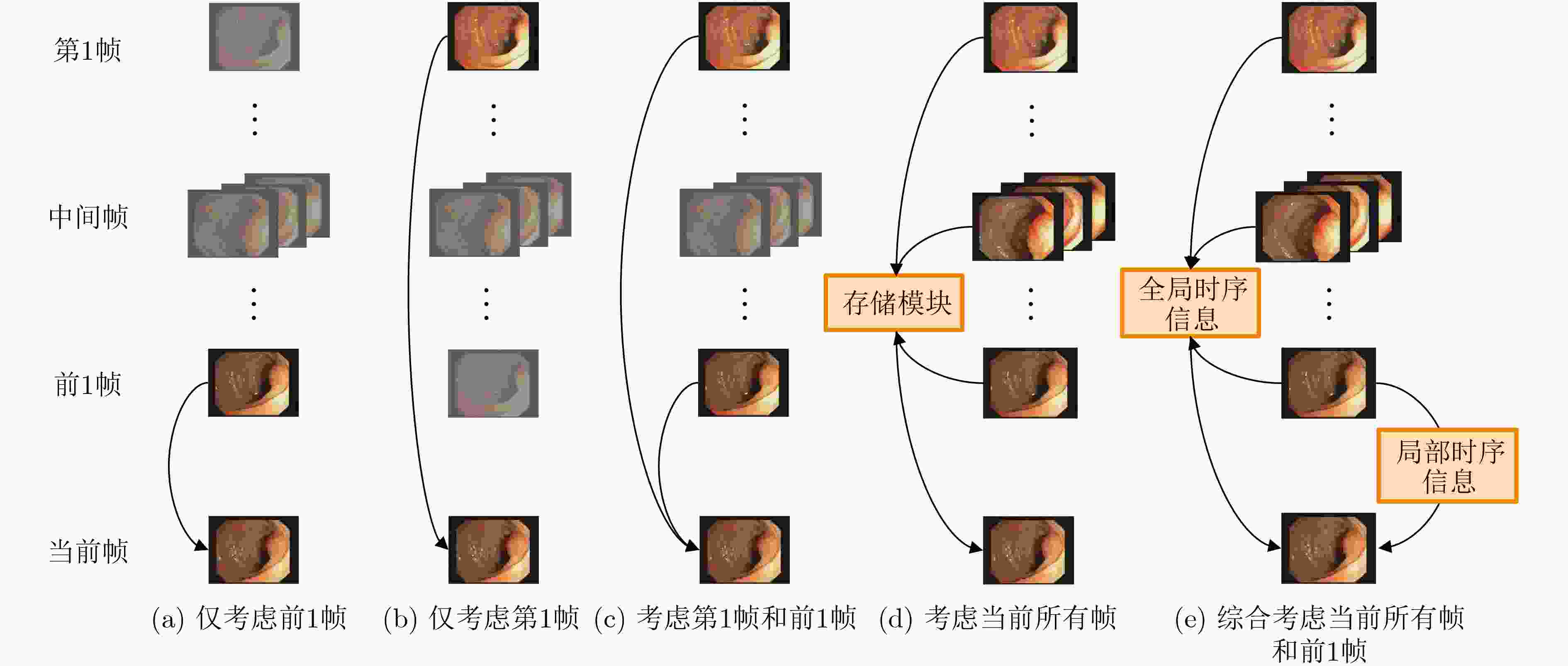
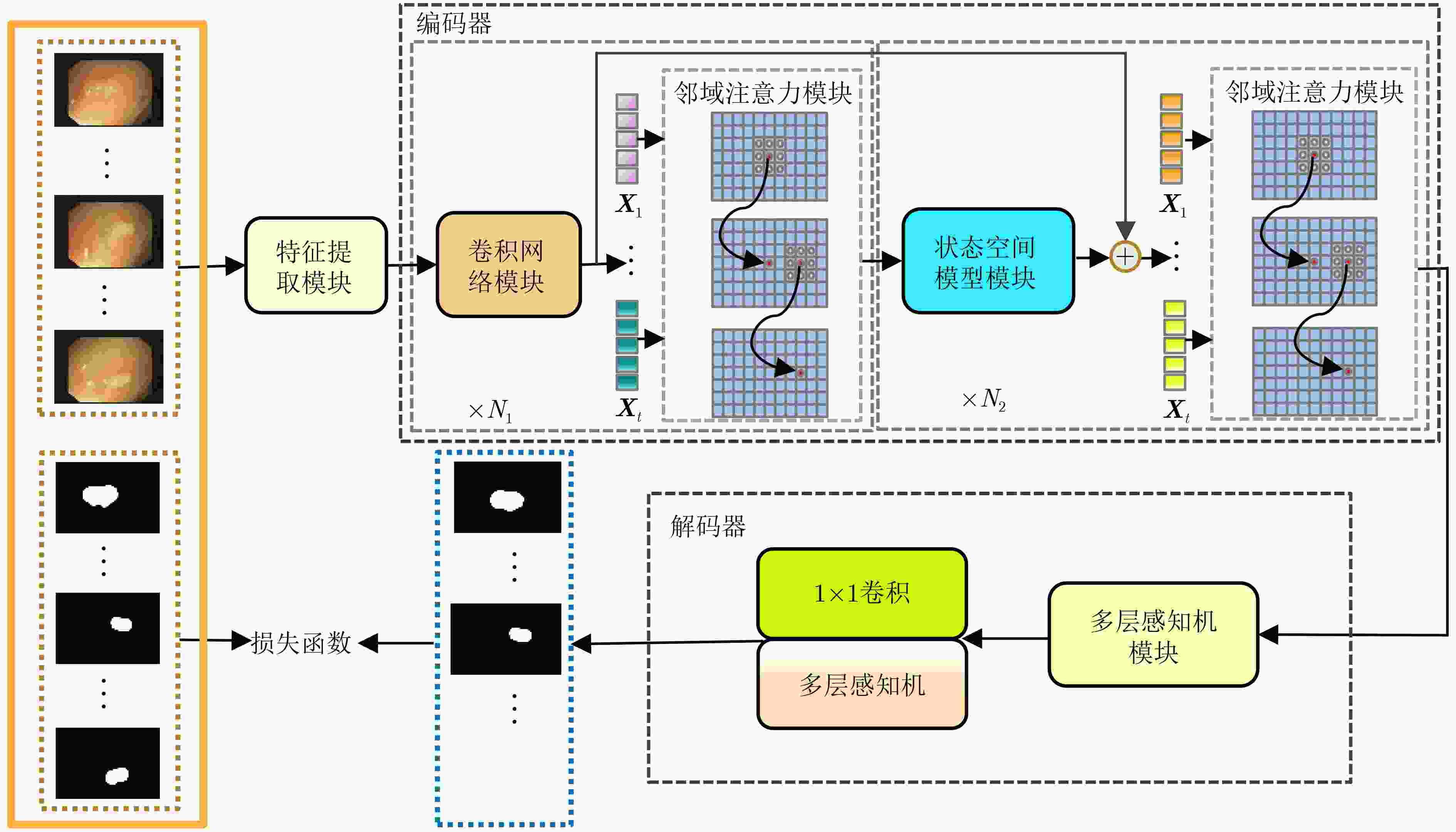
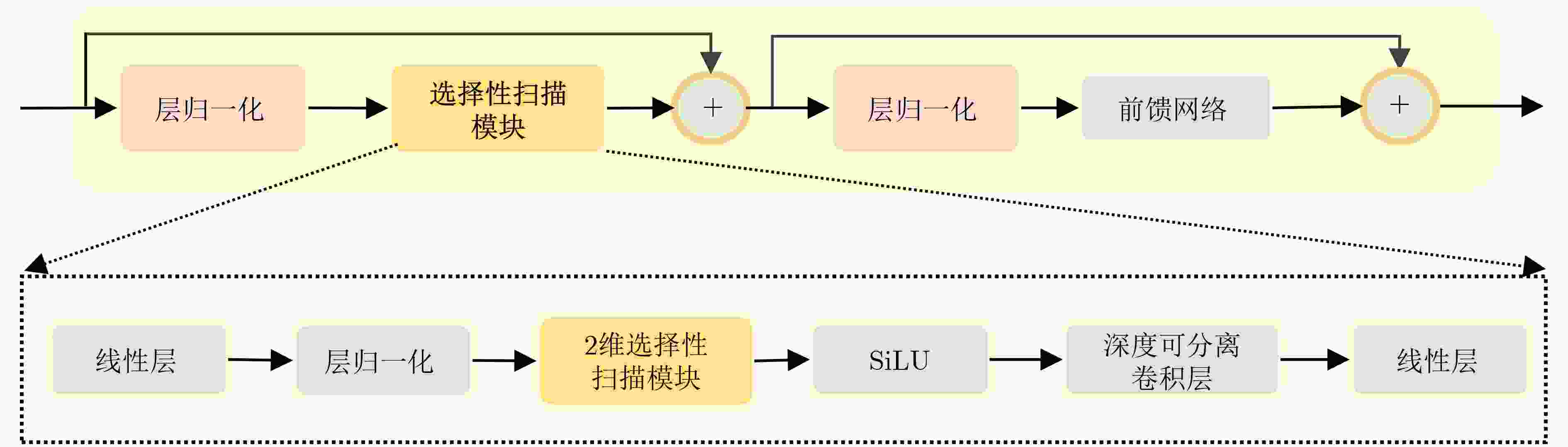
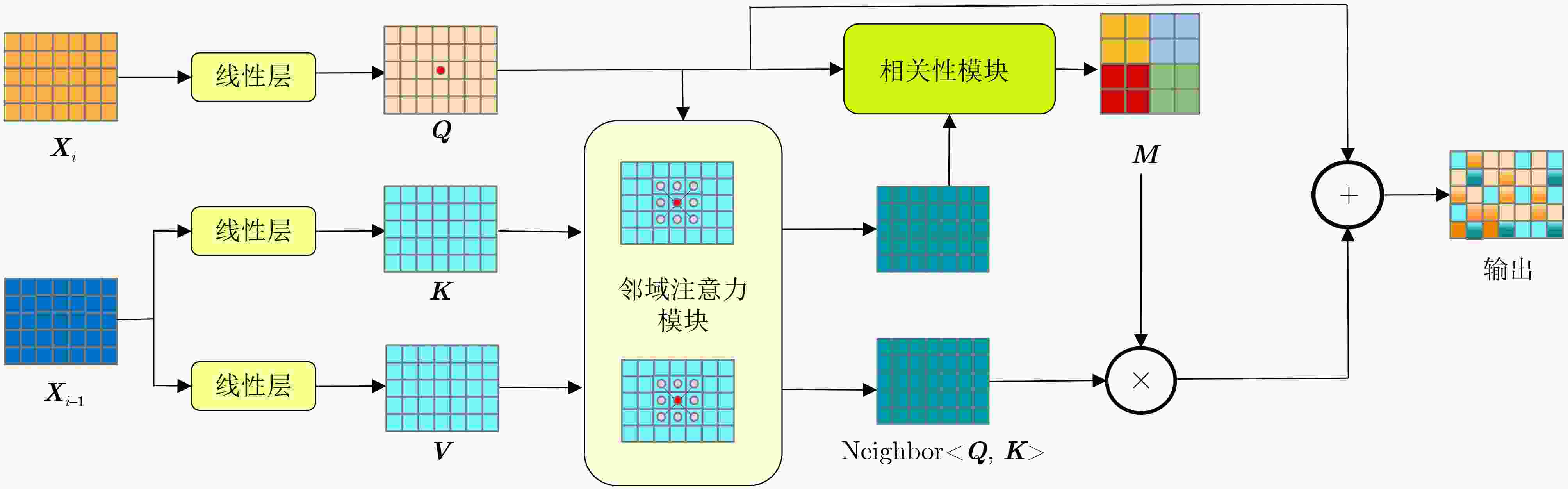
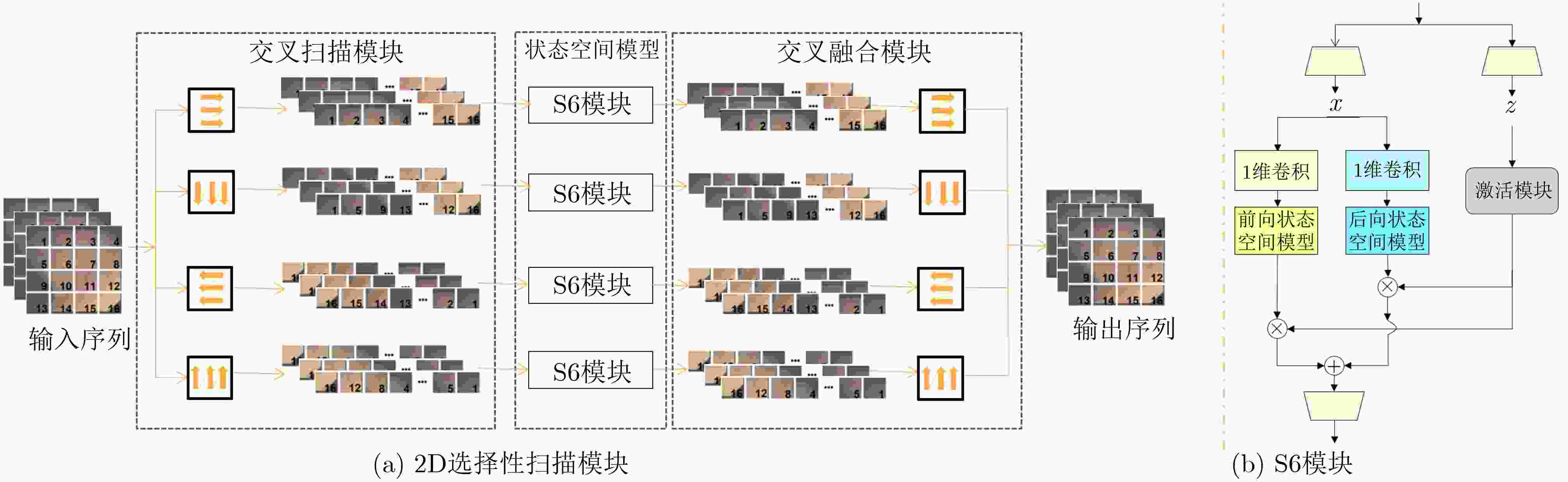
摘要: 在医学影像分析领域,精准分割视频中的病灶对于疾病的早期诊断和治疗至关重要。该文创新性地提出一种融合邻域注意力机制与状态空间模型的算法,旨在全面而精细地捕捉医学视频中的时空特征,从而对视频中的病灶进行准确分割。该算法通过两阶段的精心设计,显著提升了分割性能:第1阶段,通过深度卷积网络捕获低层次的空间语义信息,并借助邻域注意力机制,挖掘相邻帧间的局部时序语义关联。第2阶段,引入状态空间模型来捕捉全面的时序信息,并再次应用邻域注意力模块,进一步增强对局部时序特征的敏感度。该方法不仅有效整合了视频中丰富的时序信息,而且在局部和全局层面上实现了空间与时间特征的协同优化。相较于使用具有2次计算复杂度的自注意力机制,该文采用了具有线性计算复杂度的状态空间模型,显著提升了模型的训练效率和推理速度。所提算法在甲状腺超声视频数据集以及结肠息肉视频数据集CVC-ClinicDB和CVC-ColonDB上的交并比(IOU)指标分别达到了72.7%, 82.3%和72.5%,相比该文的基线模型Vivim分别提高了5.7%, 1.7%和5.5%。此外,消融实验进一步揭示了邻域注意力模块和状态空间模型在提取时序信息中发挥的关键作用。Abstract:
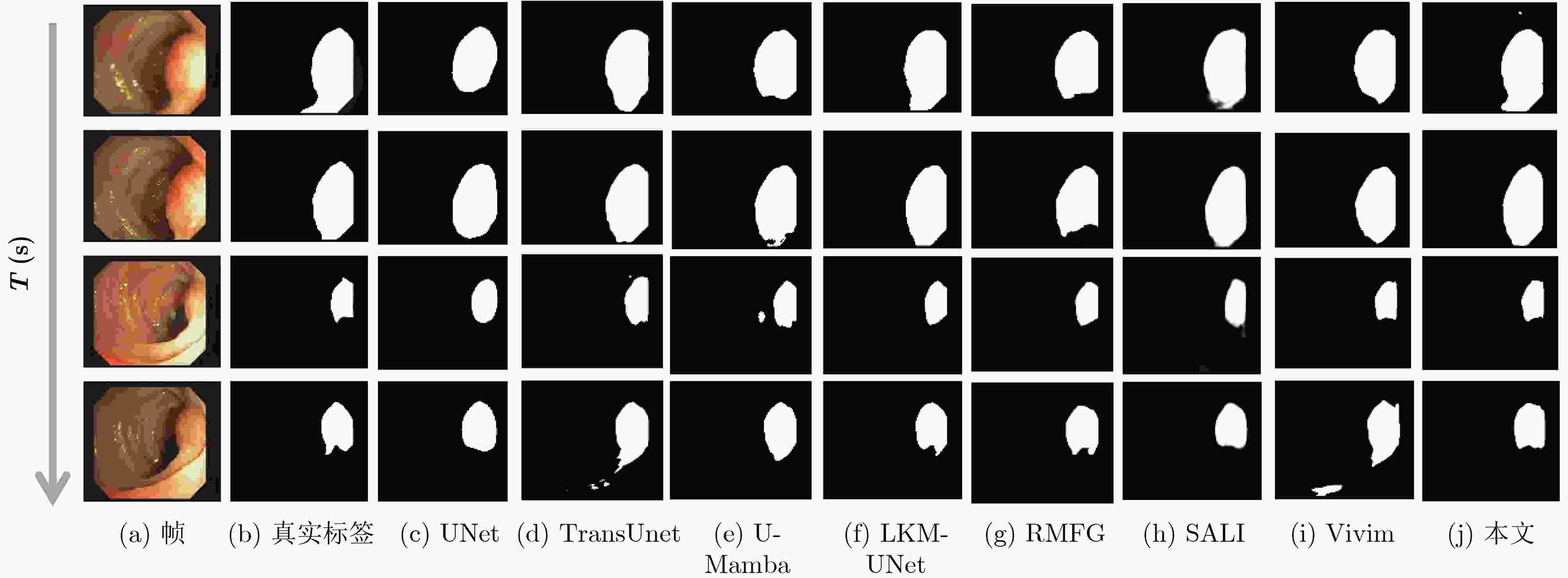
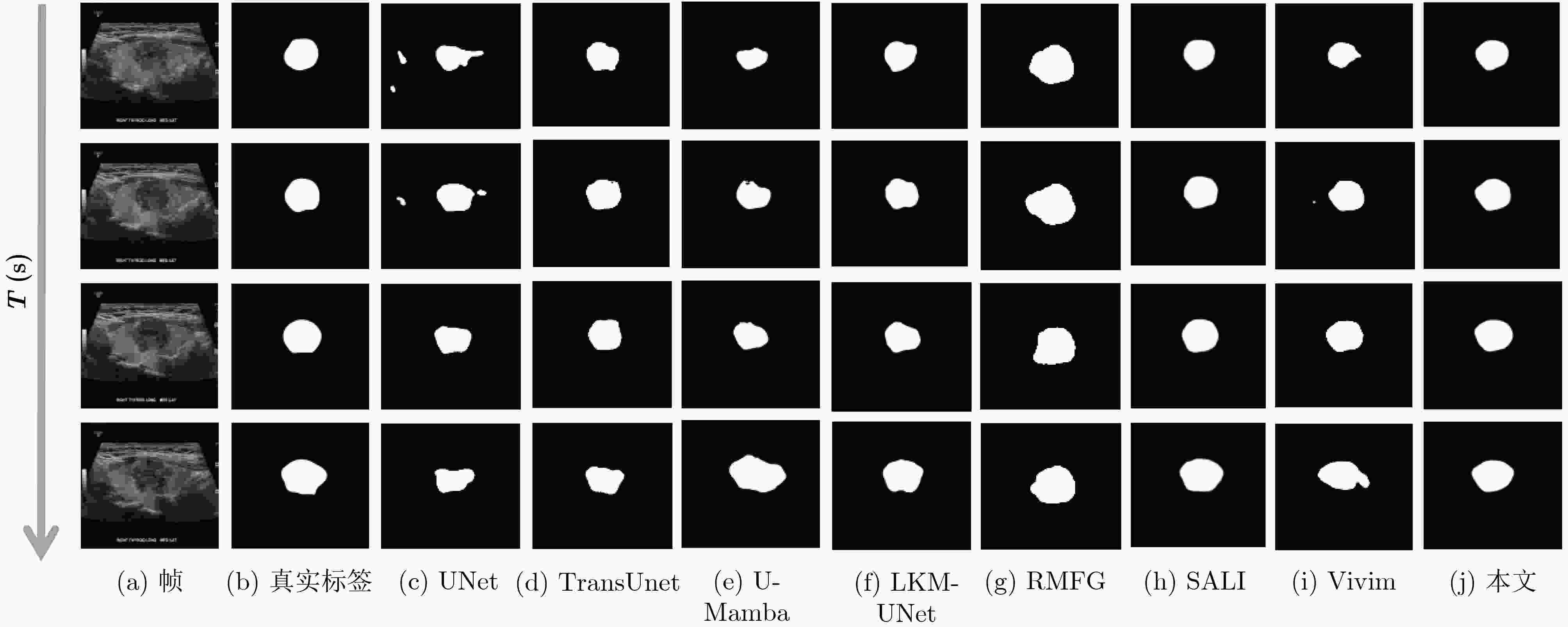
Objective Accurate segmentation of lesions in medical videos is crucial for clinical diagnosis and treatment. Unlike static medical images, videos provide continuous temporal information, enabling tracking of lesion evolution and morphological changes. However, existing segmentation methods primarily focus on processing individual frames, failing to effectively capture temporal correlations across frames. While self-attention mechanisms have been used to model long-range dependencies, their quadratic computational complexity renders them inefficient for high-resolution video segmentation. Additionally, medical videos are often affected by motion blur, noise, and illumination variations, which further hinder segmentation accuracy. To address these challenges, this paper proposes a novel medical video segmentation algorithm that integrates neighborhood attention and a State Space Model (SSM). The approach aims to efficiently capture both local and global spatiotemporal features, improving segmentation accuracy while maintaining computational efficiency. Methods The proposed approach comprises two key stages: local feature extraction and global temporal modeling, designed to efficiently capture both spatial and temporal dependencies in medical video segmentation.In the first stage, a deep convolutional network is used to extract spatial features from each video frame, providing a detailed representation of anatomical structures. However, relying solely on spatial features is insufficient for medical video segmentation, as lesions often undergo subtle morphological changes over time. To address this, a neighborhood attention mechanism is introduced to capture short-term dependencies between adjacent frames. Unlike conventional self-attention mechanisms, which compute relationships across the entire frame, neighborhood attention selectively attends to local regions around each pixel, reducing computational complexity while preserving essential temporal coherence. This localized attention mechanism enables the model to focus on small but critical changes in lesion appearance, making it more robust to motion and deformation variations. In the second stage, an SSM module is integrated to capture long-range dependencies across the video sequence. Unlike Transformer-based approaches, which suffer from quadratic complexity due to the self-attention mechanism, the SSM operates with linear complexity, significantly improving computational efficiency while maintaining strong temporal modeling capabilities. To further enhance the processing of video-based medical data, a 2D selective scanning mechanism is introduced to extend the SSM from 1D to 2D. This mechanism enables the model to extract spatiotemporal relationships more effectively by scanning input data along multiple directions and merging the results, ensuring that both local and global temporal structures are well represented. The combination of neighborhood attention for local refinement and SSM-based modeling for long-range dependencies enables the proposed method to achieve a balance between segmentation accuracy and computational efficiency. The model is trained and evaluated on multiple medical video datasets to verify its effectiveness across different segmentation scenarios, demonstrating its capability to handle complex lesion appearances, background noise, and variations in imaging conditions. Results and Discussions The proposed method is evaluated on three widely used medical video datasets: thyroid ultrasound, CVC-ClinicDB, and CVC-ColonDB. The model achieves Intersection Over Union (IOU) scores of 72.7%, 82.3%, and 72.5%, respectively, outperforming existing state-of-the-art methods. Compared to the Vivim model, the proposed method improves IOU by 5.7%, 1.7%, and 5.5%, highlighting the advantage of leveraging temporal information. In terms of computational efficiency, the model achieves 23.97 frames per second (fps) on the thyroid ultrasound dataset, making it suitable for real-time clinical applications. A comparative analysis against several state-of-the-art methods, including UNet, TransUNet, PraNet, U-Mamba, LKM-UNET, RMFG, SALI, and Vivim, demonstrates that the proposed method consistently outperforms these approaches, particularly in complex scenarios with significant background noise, occlusions, and motion artifacts. Specifically, on the CVC-ClinicDB dataset, the proposed model achieves an IOU of 82.3%, exceeding the previous best approach (80.9%). On the CVC-ColonDB dataset, which presents additional challenges due to lighting variations and occlusions, the model attains an IOU of 72.5%, outperforming the previous best method (70.8%). These results highlight the importance of incorporating both local and global temporal information to enhance segmentation accuracy and robustness in medical video analysis. Conclusions This study proposes a medical video segmentation algorithm that integrates neighborhood attention and an SSM to capture both local and global spatiotemporal features. This integration enables an effective balance between segmentation accuracy and computational efficiency. Experimental results demonstrate the superiority of the proposed method over existing approaches across multiple medical video datasets. The main contributions include: the combined use of neighborhood attention and SSM for efficient spatiotemporal feature extraction; a 2D selective scanning mechanism that extends SSMs for video-based medical segmentation; improved segmentation performance exceeding that of state-of-the-art models while maintaining real-time processing capability; and enhanced robustness to background noise and lighting variations, improving reliability in clinical applications. Future work will focus on incorporating prior knowledge and anatomical constraints to refine segmentation accuracy in cases with ambiguous lesion boundaries; developing advanced boundary refinement strategies for challenging scenarios; extending the framework to multi-modal imaging data such as CT and MRI videos; and optimizing the model for deployment on edge devices to support real-time processing in point-of-care and mobile healthcare settings. -
表 1 甲状腺超声数据集实验结果
方法 对象 IOU↑ Dice↑ Precision↑ Recall↑ MAE↓ GFLOPS↓ (G) Params↓ (M) fps↑ UNet image 0.607 0.718 0.885 0.628 0.017 65.52 34.53 54.87 UNeXt image 0.635 0.766 0.875 0.703 0.018 0.57 1.47 66.14 TransUnet image 0.629 0.739 0.775 0.758 0.022 32.23 93.23 46.47 PraNet image 0.642 0.752 0.846 0.697 0.028 6.96 30.50 6.96 U-Mamba image 0.648 0.749 0.845 0.694 0.016 58.18 76.38 30.86 LKM-UNet image 0.656 0.792 0.702 0.909 0.020 104.31 189.53 20.73 RMFG video 0.689 0.777 0.834 0.735 0.021 6.28 27.87 5.71 SALI video 0.681 0.795 0.707 0.951 0.014 69.27 82.73 4.77 Vivim video 0.688 0.791 0.833 0.785 0.034 19.18 53.63 26.67 本文 video 0.727 0.829 0.903 0.801 0.013 19.71 56.53 23.97 表 2 结肠数据集实验结果
数据集 评价指标 UNet UNeXt TransUnet PraNet U-Mamba LKM-UNET RMFG SALI Vivim 本文 CVC-
ClinicDBIOU↑
Dice↑
Precision↑
Recall↑
MAE↓0.607
0.750
0.993
0.613
0.0980.705
0.807
0.895
0.784
0.0440.714
0.825
0.779
0.899
0.0490.783
0.875
0.802
0.972
0.0380.766
0.861
0.771
0.993
0.0150.777
0.874
0.995
0.780
0.0520.798
0.883
0.920
0.862
0.0340.785
0.880
0.832
0.933
0.0420.809
0.883
0.913
0.880
0.0170.823
0.889
0.925
0.876
0.014CVC-
ColonDBIOU↑
Dice↑
Precision↑
Recall↑
MAE↓0.593
0.731
0.812
0.729
0.0140.555
0.662
0.764
0.670
0.0750.658
0.736
0.871
0.728
0.0580.666
0.777
0.752
0.895
0.0300.694
0.796
0.702
0.980
0.0150.705
0.824
0.711
0.988
0.0270.659
0.791
0.713
0.931
0.0140.708
0.829
0.719
0.978
0.0340.687
0.805
0.955
0.718
0.0200.725
0.834
0.972
0.743
0.017表 3 消融实验结果表
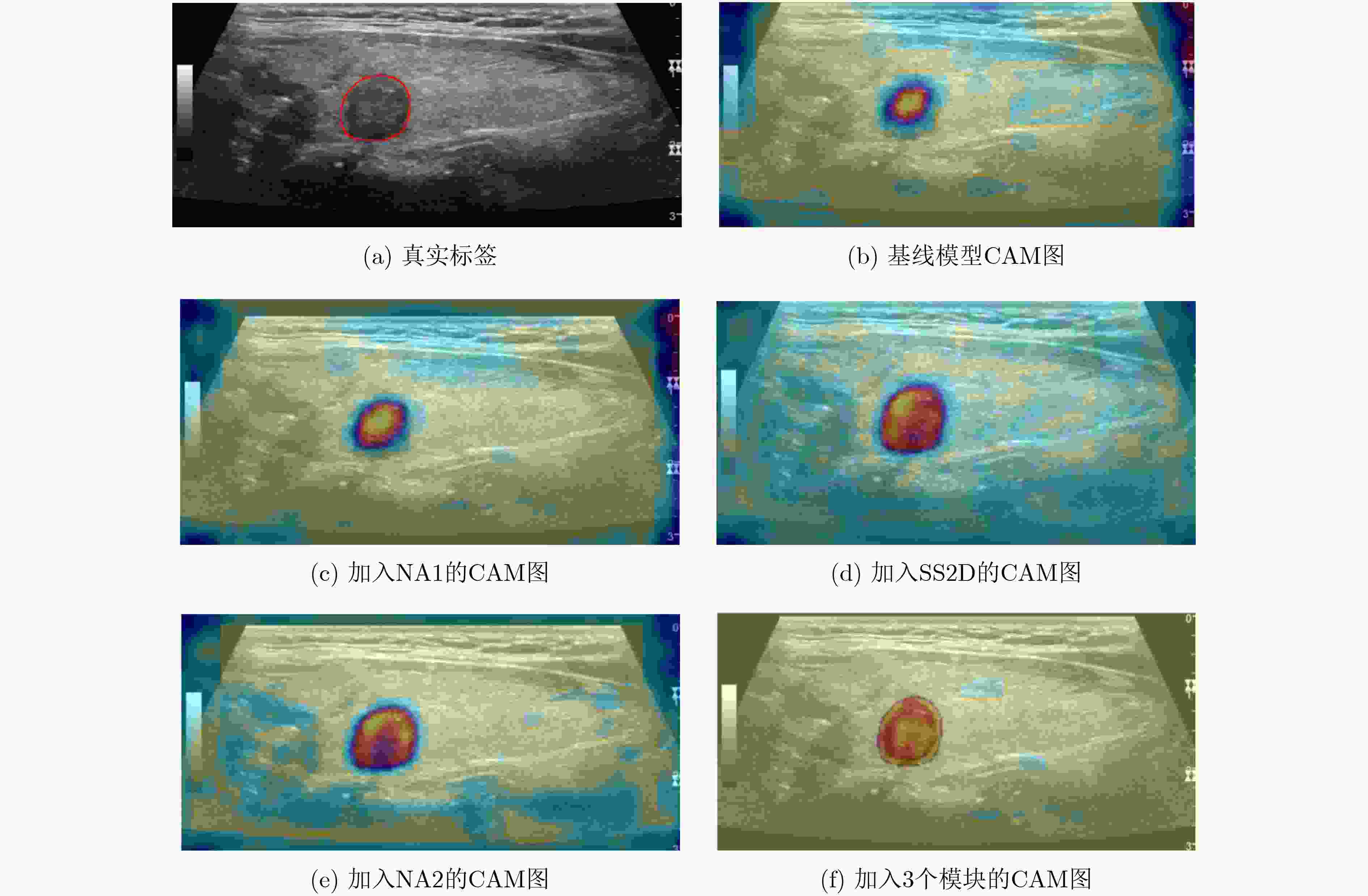
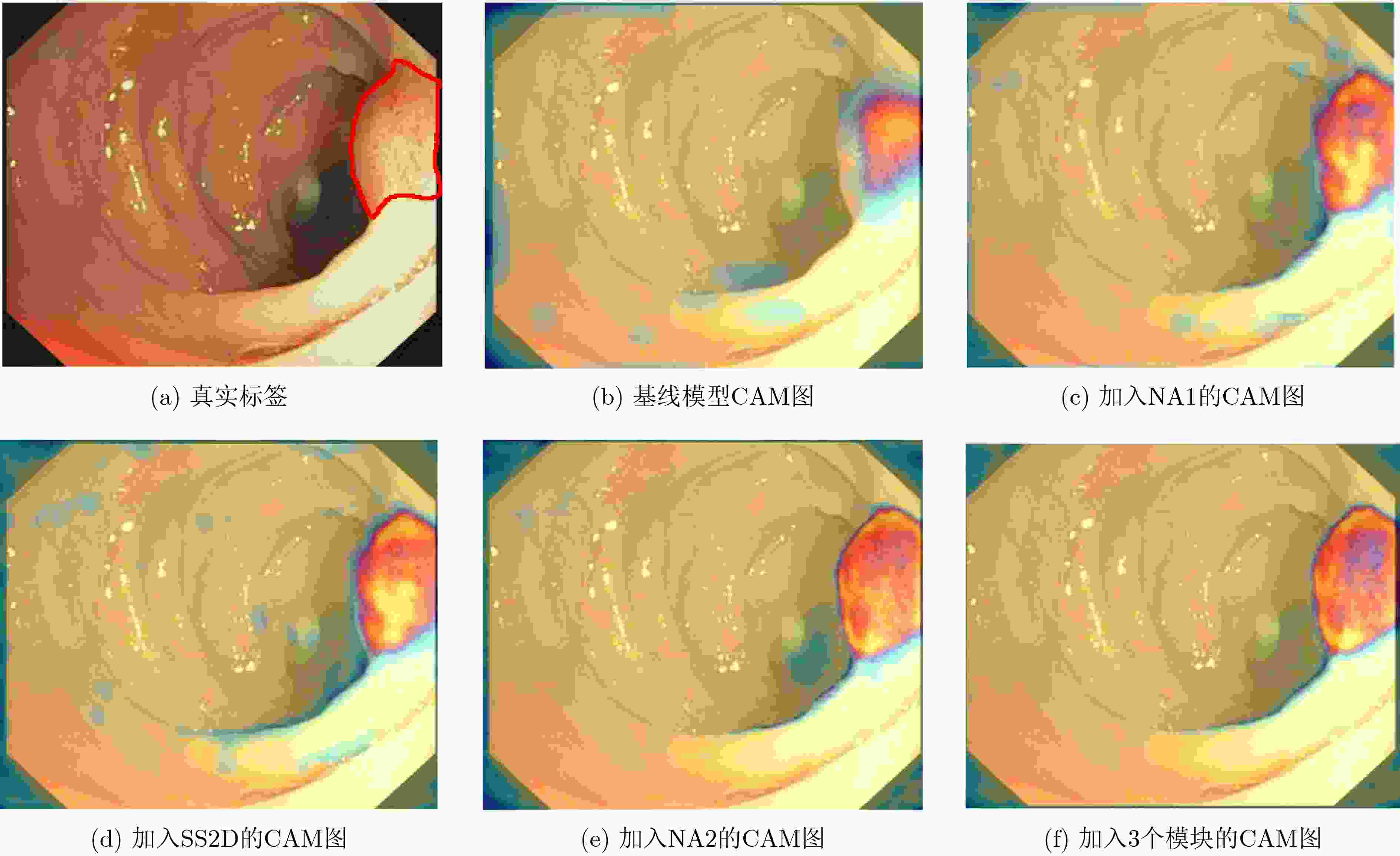
数据集 基线
模型邻域注意力
模块12维选择性
扫描模块邻域注意力
模块2IOU↑ Dice↑ Precision↑ Recall↑ MAE↓ 甲状腺
数据集√ 0.688 0.791 0.833 0.785 0.034 √ √ 0.707 0.809 0.902 0.771 0.015 √ √ 0.716 0.821 0.900 0.786 0.014 √ √ 0.703 0.813 0.904 0.773 0.015 √ √ √ √ 0.727 0.829 0.903 0.801 0.013 CVC-ClinicDB √ 0.809 0.883 0.913 0.880 0.017 √ √ 0.814 0.885 0.928 0.870 0.017 √ √ 0.818 0.887 0.926 0.875 0.016 √ √ 0.816 0.886 0.926 0.873 0.016 √ √ √ √ 0.823 0.889 0.925 0.876 0.014 -
[1] MINAEE S, BOYKOV Y, PORIKLI F, et al. Image segmentation using deep learning: A survey[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022, 44(7): 3523–3542. doi: 10.1109/TPAMI.2021.3059968. [2] GHOSH S, DAS N, DAS I, et al. Understanding deep learning techniques for image segmentation[J]. ACM Computing Surveys, 2020, 52(4): 73. doi: 10.1145/3329784. [3] MA Jun, HE Yuting, LI Feifei, et al. Segment anything in medical images[J]. Nature Communications, 2024, 15(1): 654. doi: 10.1038/s41467-024-44824-z. [4] PATIL D D and DEORE S G. Medical image segmentation: A review[J]. International Journal of Computer Science and Mobile Computing, 2013, 2(1): 22–27. [5] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. 18th International Conference on Medical Image Computing and Computer Assisted Intervention, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [6] VALANARASU J M J and PATEL V M. UNeXt: MLP-based rapid medical image segmentation network[C]. 25th International Conference on Medical Image Computing and Computer Assisted Intervention, Singapore, 2022: 23–33. doi: 10.1007/978-3-031-16443-9_3. [7] VASWANI A, SHAZEER N, PARMAR N, et al. Attention is all you need[C]. Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, USA, 2017: 6000–601. [8] DOSOVITSKIY A, BEYER L, KOLESNIKOV A, et al. An image is worth 16x16 words: Transformers for image recognition at scale[C]. 9th International Conference on Learning Representations, Austria, 2021. [9] CHEN Jieneng, LU Yongyi, YU Qihang, et al. TransUNet: Transformers make strong encoders for medical image segmentation[Z]. arXiv: 2102.04306, 2021. doi: 10.48550/arXiv.2102.04306. [10] CHOROMANSKI K M, LIKHOSHERSTOV V, DOHAN D, et al. Rethinking attention with performers[C]. 9th International Conference on Learning Representations, Austria, 2021. [11] KITAEV N, KAISER Ł, and LEVSKAYA A. Reformer: The efficient transformer[C]. 8th International Conference on Learning Representations, Addis Ababa, Ethiopia, 2020. [12] QIN Zhen, YANG Songlin, and ZHONG Yiran. Hierarchically gated recurrent neural network for sequence modeling[C]. The 37th International Conference on Neural Information Processing Systems, New Orleans, USA, 2023: 1442. [13] BASAR T. A new approach to linear filtering and prediction problems[M]. BASAR T. Control Theory: Twenty-Five Seminal Papers (1932–1981). Wiley-IEEE Press, 2001: 167–179. doi: 10.1109/9780470544334.ch9. [14] GU A and DAO T. Mamba: Linear-time sequence modeling with selective state spaces[Z]. arXiv: 2312.00752, 2023. doi: 10.48550/arXiv.2312.00752. [15] GU A, GOEL K, and RÉ C. Efficiently modeling long sequences with structured state spaces[C]. 10th International Conference on Learning Representations, 2022. [16] GU A, JOHNSON I, GOEL K, et al. Combining recurrent, convolutional, and continuous-time models with linear state-space layers[C]. The 35th International Conference on Neural Information Processing Systems, 2021: 44. [17] ZHU Lianghui, LIAO Bencheng, ZHANG Qian, et al. Vision mamba: Efficient visual representation learning with bidirectional state space model[C]. The Forty-first International Conference on Machine Learning, Vienna, Austria, 2024. [18] NGUYEN E, GOEL K, GU A, et al. S4ND: Modeling images and videos as multidimensional signals using state spaces[C]. The 36th International Conference on Neural Information Processing Systems, New Orleans, USA, 2022: 206. [19] LIU Yue, TIAN Yunjie, ZHAO Yuzhong, et al. VMamba: Visual state space model[C]. The 38th Conference on Neural Information Processing Systems, Vancouver, Canada, 2024. [20] MA Jun, LI Feifei, and WANG Bo. U-Mamba: Enhancing long-range dependency for biomedical image segmentation[Z]. arXiv: 2401.04722, 2024. doi: 10.48550/arXiv.2401.04722. [21] XING Zhaohu, YE Tian, YANG Yijun, et al. SegMamba: Long-range sequential modeling mamba for 3D medical image segmentation[C]. 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 578–588. doi: 10.1007/978-3-031-72111-3_54. [22] WANG Jinhong, CHEN Jintai, CHEN D, et al. LKM-UNet: Large kernel vision mamba UNet for medical image segmentation[C]. 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 360–370. doi: 10.1007/978-3-031-72111-3_34. [23] XU Zhongxing, TANG Feilong, CHEN Zhe, et al. Polyp-Mamba: Polyp segmentation with visual mamba[C]. 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 510–521. doi: 10.1007/978-3-031-72111-3_48. [24] ARNAB A, DEHGHANI M, HEIGOLD G, et al. ViViT: A video vision transformer[C]. The IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 6816–6826. doi: 10.1109/ICCV48922.2021.00676. [25] ZHANG Miao, LIU Jie, WANG Yifei, et al. Dynamic context-sensitive filtering network for video salient object detection[C]. The IEEE/CVF International Conference on Computer Vision, Montreal, Canada, 2021: 1533–1543. doi: 10.1109/ICCV48922.2021.00158. [26] LI Jialu, ZHENG Qingqing, LI Mingshuang, et al. Rethinking breast lesion segmentation in ultrasound: A new video dataset and a baseline network[C]. 25th International Conference on Medical Image Computing and Computer Assisted Intervention, Singapore, Singapore, 2022: 391–400. doi: 10.1007/978-3-031-16440-8_38. [27] LIN Junhao, DAI Qian, ZHU Lei, et al. Shifting more attention to breast lesion segmentation in ultrasound videos[C]. 26th International Conference on Medical Image Computing and Computer Assisted Intervention, Vancouver, Canada, 2023: 497–507. doi: 10.1007/978-3-031-43898-1_48. [28] HU Xifeng, CAO Yankun, HU Weifeng, et al. Refined feature-based Multi-frame and Multi-scale Fusing Gate network for accurate segmentation of plaques in ultrasound videos[J]. Computers in Biology and Medicine, 2023, 163: 107091. doi: 10.1016/j.compbiomed.2023.107091. [29] SONG Qi, LI Jie, LI Chenghong, et al. Fully attentional network for semantic segmentation[C]. The 36th AAAI Conference on Artificial Intelligence, Washington, USA, 2022: 2280–2288. doi: 10.1609/aaai.v36i2.20126. [30] DOSOVITSKIY A, FISCHER P, ILG E, et al. FlowNet: Learning optical flow with convolutional networks[C]. The IEEE International Conference on Computer Vision, Santiago, Chile, 2015: 2758–2766. doi: 10.1109/ICCV.2015.316. [31] JI Gepeng, CHOU Yucheng, FAN Dengping, et al. Progressively normalized self-attention network for video polyp segmentation[C]. 24th International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 2021: 142–152. doi: 10.1007/978-3-030-87193-2_14. [32] HU Qiang, YI Zhenyu, ZHOU Ying, et al. SALI: Short-term alignment and long-term interaction network for colonoscopy video polyp segmentation[C]. 27th International Conference on Medical Image Computing and Computer Assisted Intervention, Marrakesh, Morocco, 2024: 531–541. doi: 10.1007/978-3-031-72089-5_50. [33] HU Yuanting, HUANG Jiabin, and SCHWING A G. MaskRNN: Instance level video object segmentation[C]. The 31st International Conference on Neural Information Processing Systems, Long Beach, USA, 2017: 324–333. [34] HU Yuanting, HUANG Jiabin, and SCHWING A G. VideoMatch: Matching based video object segmentation[C]. 15th European Conference on Computer Vision, Munich, Germany, 2018: 56–73. doi: 10.1007/978-3-030-01237-3_4. [35] Seoung W O, LEE J Y, SUNKAVALLI K, et al. Fast video object segmentation by reference-guided mask propagation[C]. The IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 7376–7385. doi: 10.1109/CVPR.2018.00770. [36] Seoung W O, LEE J Y, XU Ning, et al. Video object segmentation using space-time memory networks[C]. Tthe IEEE/CVF International Conference on Computer Vision, Seoul, Korea (South), 2019: 9225–9234. doi: 10.1109/ICCV.2019.00932. [37] YANG Yijun, XING Zhaohu, and ZHU Lei. Vivim: A video vision mamba for medical video object segmentation[Z]. arXiv: 2401.14168, 2024. doi: 10.48550/arXiv.2401.14168. [38] HASSANI A, WALTON S, LI Jiachen, et al. Neighborhood attention transformer[C]. The IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, Canada, 2023: 6185–6194. doi: 10.1109/CVPR52729.2023.00599. [39] WANG Xiaolong, GIRSHICK R, GUPTA A, et al. Non-local neural networks[C]. The IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, USA, 2018: 7794–7803. doi: 10.1109/CVPR.2018.00813. [40] PENG Chao, ZHANG Xiangyu, YU Gang, et al. Large kernel matters — Improve semantic segmentation by global convolutional network[C]. The IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, USA, 2017: 1743–1751. doi: 10.1109/CVPR.2017.189. [41] BERNAL J, SÁNCHEZ F J, FERNÁNDEZ-ESPARRACH G, et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. Saliency maps from physicians[J]. Computerized Medical Imaging and Graphics, 2015, 43: 99–111. doi: 10.1016/j.compmedimag.2015.02.007. [42] BERNAL J, SÁNCHEZ J, and VILARIÑO F. Towards automatic polyp detection with a polyp appearance model[J]. Pattern Recognition, 2012, 45(9): 3166–3182. doi: 10.1016/j.patcog.2012.03.002. [43] Stanford AIMI. AIMI dataset[EB/OL]. https://stanfordaimi.azurewebsites.net/datasets/a72f2b02–7b53–4c5d-963c-d7253220bfd5, 2021. [44] WANG Yi, DENG Zijun, HU Xiaowei, et al. Deep attentional features for prostate segmentation in ultrasound[C]. 21st International Conference on Medical Image Computing and Computer Assisted Intervention, Granada, Spain, 2018: 523–530. doi: 10.1007/978-3-030-00937-3_60. [45] FAN Dengping, JI Gepeng, ZHOU Tao, et al. PraNet: Parallel reverse attention network for polyp segmentation[C]. 23rd International Conference on Medical Image Computing and Computer Assisted Intervention, Lima, Peru, 2020: 263–273. doi: 10.1007/978-3-030-59725-2_26. [46] MA Ningning, ZHANG Xiangyu, ZHENG Haitao, et al. ShuffleNet V2: Practical guidelines for efficient CNN architecture design[C]. The 15th European Conference on Computer Vision, Munich, Germany, 2018: 122–138. doi: 10.1007/978-3-030-01264-9_8. [47] SELVARAJU R R, COGSWELL M, DAS A, et al. Grad-CAM: Visual explanations from deep networks via gradient-based localization[C]. IEEE International Conference on Computer Vision, Venice, Italy, 2017: 618–626. doi: 10.1109/ICCV.2017.74. -





 下载:
下载:


 下载:
下载:
