Efficient File Random Access Method Based on Primer Index Matrix in DNA Storage Scenarios
-
摘要: DNA分子具有密度高和稳定性的优势,有望成为下一代海量数据存储需求的介质,近年来受到广泛关注。目前将引物作为文件的唯一标识,基于聚合酶链式反应(PCR)扩增技术可实现对DNA池存储文件的随机检索,但对引物与文件之间的分配和映射关系没有进行深入研究,仍然采用随机分配的方式来关联引物与文件,这会导致目标引物序列的查找效率降低,且保存引物与文件的映射关系表会造成大量的数据冗余。为了提供一种高效的硅基计算设备与碳基存储系统的连接桥梁,有效降低存储引物与文件映射关系所带来的数据冗余,该文提出一种基于引物索引矩阵的DNA存储随机检索方法。该方法通过将存储文件集按照文件的不同属性进行划分来构建引物索引矩阵,同时将引物库中的引物按照转换规则转化为有序引物库,最后优化引物与文件之间的映射关系,以实现对文件的高效、多维度检索。实验结果表明,在存储不同规模的文件集时,运用所提算法建立对应的引物索引矩阵,可将引物检索效率提高为常数级时间复杂度,并且存储引物与文件的映射关系所需要的额外存储空间从原来的线性增长优化为对数增长。Abstract: DNA molecules have the advantages of high density and stability, and are expected to become the medium for the next generation of massive data storage needs, which has received widespread attention in recent years. Currently, primers are used as the unique identifier for files, and random retrieval of DNA pool storage files can be achieved based on polymerase chain reaction (PCR) amplification technology. However, the allocation and mapping relationship between primers and files have not been thoroughly studied, and random allocation is still used to associate primers and files. This will lead to a decrease in the search efficiency of the target primer sequence, and saving the mapping relationship table between primers and files will cause a lot of data redundancy. In order to provide an efficient connection bridge between silicon-based computing devices and carbon-based storage systems, and effectively reduce the data redundancy caused by storing primer-file mapping relationships, a random retrieval method for DNA storage based on the primer index matrix is proposed in this paper. This method constructs a primer index matrix by dividing the stored file set according to different attributes of the file, and converts the primers in the primer library into an ordered primer library according to conversion rules. Finally, the mapping relationship between primers and files is optimized to achieve efficient and multi-dimensional retrieval during file random retrieval. The experimental results show that when storing file sets of different sizes, the efficiency of primer retrieval is improved to a constant level of time complexity by establishing the corresponding primer index matrix using the proposed algorithm in this paper, and the extra storage space required to store the mapping relationship between primers and files is optimized from linear growth to logarithmic growth.
-
Key words:
- DNA storage /
- Random search /
- Primer index matrix
-
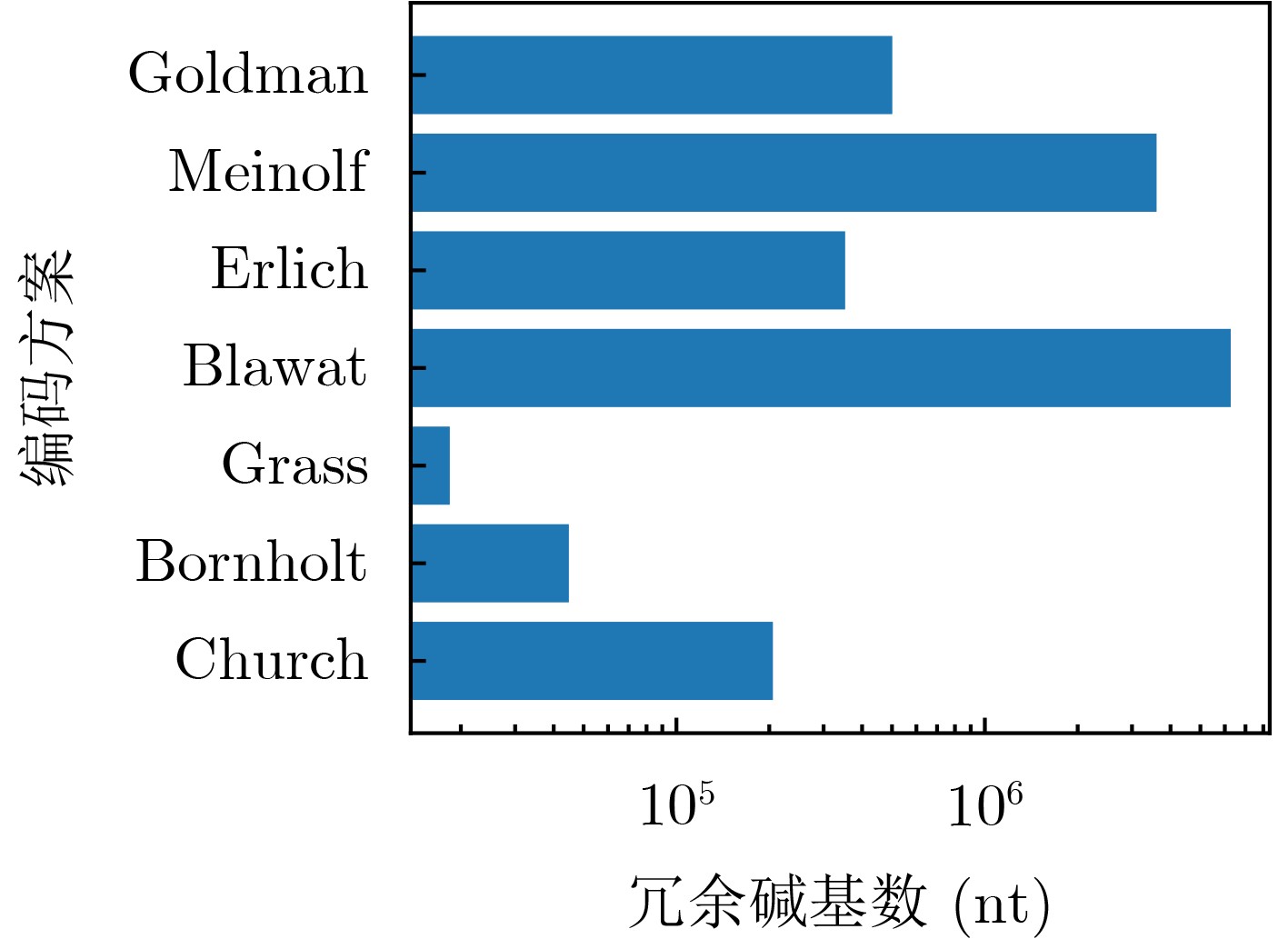
表 1 空间节省率实验测试参数表
方案 数据存储量(MB) 信息密度(bit/nt) 单池碱基数(个) 引物库大小(条) 引物库信息占用空间(Byte) Church等人 0.65 0.83 $6.42 \times {10^6}$ 666 $2.13 \times {10^4}$ Bornholt等人 0.15 0.88 $1.40 \times {10^6}$ 154 $0.49 \times {10^4}$ Grass等人 0.08 1.14 $0.57 \times {10^6}$ 82 $0.26 \times {10^4}$ Blawat等人 22.00 0.92 $1.96 \times {10^8}$ 22528 $7.21 \times {10^5}$ Erlich等人 2.11 1.57 $1.10 \times {10^7}$ 2161 $6.92 \times {10^4}$ Meinolf等人 22.00 1.60 $1.13 \times {10^8}$ 22528 $ 7.21 \times {10^5} $ Goldman等人 0.63 0.33 $1.57 \times {10^7}$ 646 $2.07 \times {10^4}$ 表 2 实验设备配置清单
设备类型 配置信息 处理器 Intel(R) Core(TM) i5-7300HQ CPU @ 2.50 GHz RAM 16.0 GB 位数 64 位 操作系统
硬盘Windows 10
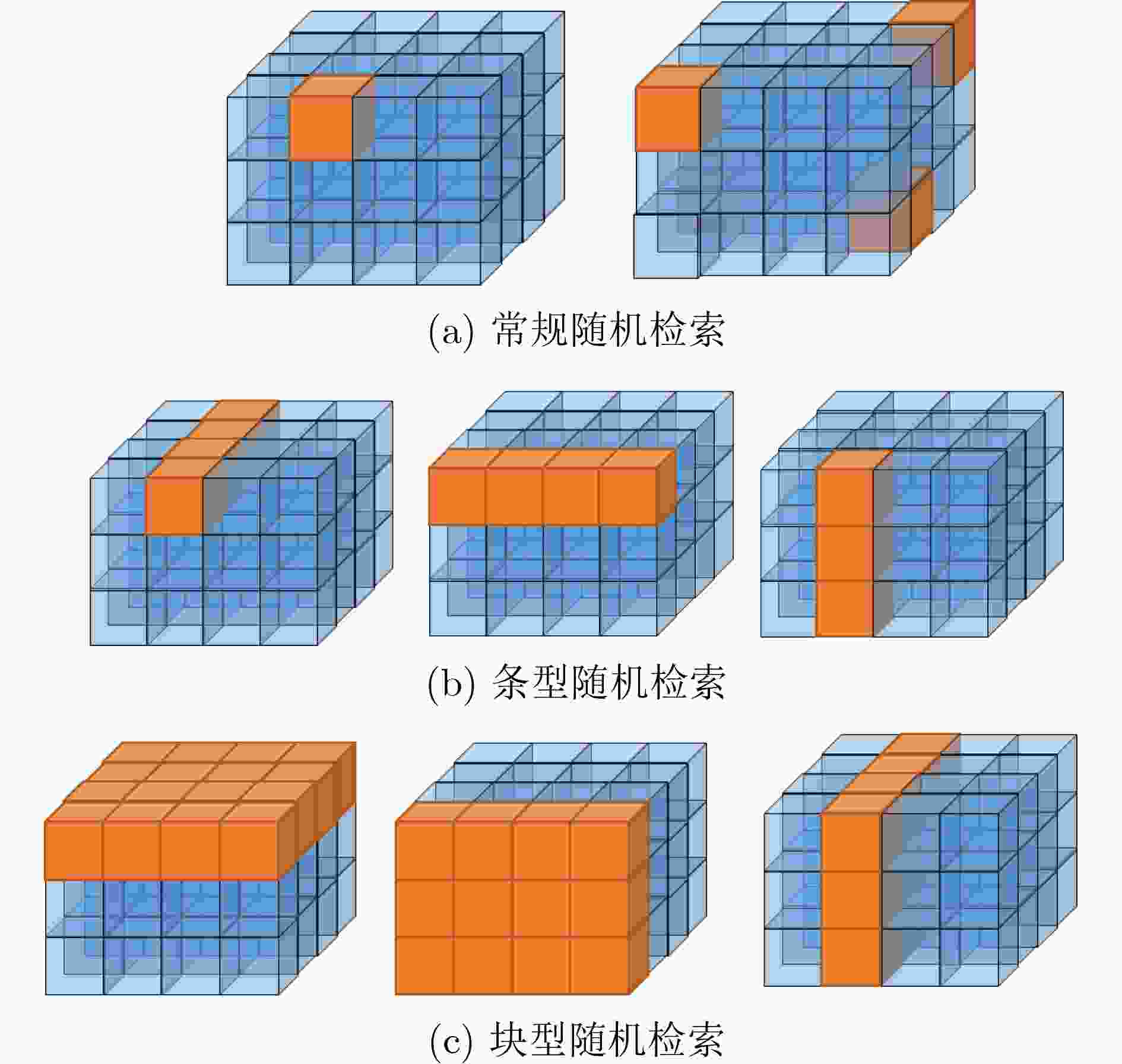
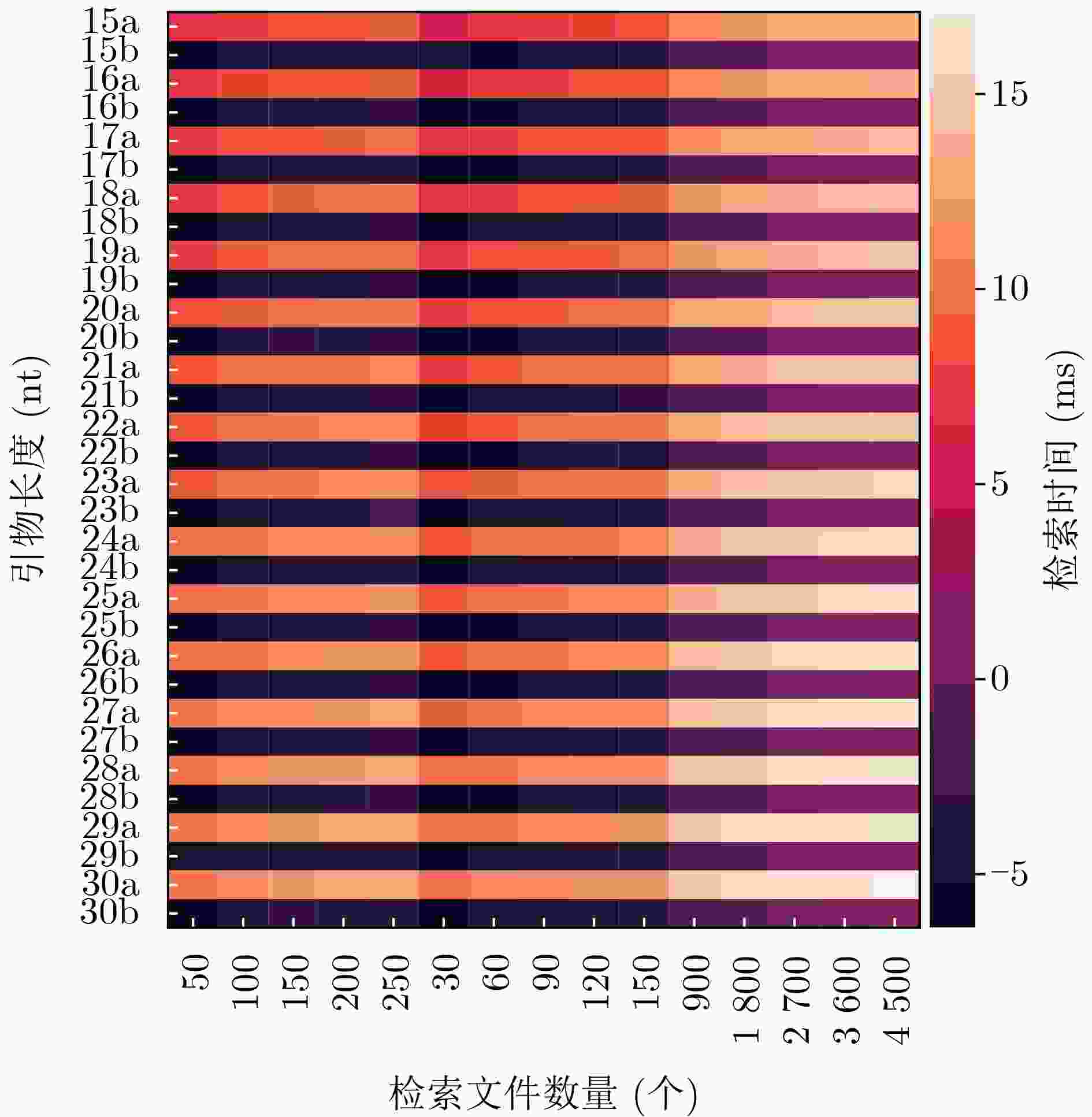
Samsung SSD 980 500 GB表 3 不同随机检索模式的检索效率
引物长度N(nt) 引物库大小 引物索引矩阵形状三元组
(R,C,L)检索模式 检索文件数量 传统方法平均检索
时长t(ms)本文方法平均检索
时长t(ms)15 1000 (10,10,10) 常规 1 0.72 0.01 15
15
15
15
15
15
15
30
30
30
30
30
30
30
301000
1000
1000
32000
32000
32000
32000
1000
1000
1000
1000
32000
32000
32000
32000(10,10,10)
(10,10,10)
(10,10,10)
(40,40,20)
(40,40,20)
(40,40,20)
(40,40,20)
(10,10,10)
(10,10,10)
(10,10,10)
(10,10,10)
(40,40,20)
(40,40,20)
(40,40,20)
(40,40,20)常规
条型
块型
常规
常规
条型
块型
常规
常规
条型
块型
常规
常规
条型
块型500
10
100
1
500
40
1600
1
500
10
100
1
500
40
160017.20
1.07
4.00
23.68
576.75
69.52
1836.68
0.80
16.76
1.40
4.28
24.32
666.02
70.02
2134.152.25
0.02
0.87
0.01
2.41
0.17
9.08
0.01
2.19
0.01
1.20
0.01
2.09
0.20
10.12表 4 不同方案的性能比较
设计方案 文件-引物映射关系 文件数:引物 有效载荷占比 多模式检索 检索复杂度 引物库信息复用 Organick等人 一对一 1:1 0.6 不支持 $O(n)$ 不支持 Song等人 多对一 $n:{\log _4}n$ 0.2 支持 $O({\log _4}n)$ 不支持 本文 一对一 1:1 0.6 支持 $O(1)$ 支持 -
[1] WIENER N. Machines smarter than men? Interview with Dr. Norbert wiener, noted scientist[R]. 1964: 84–86. [2] WELTER L, MAAROUF I, LENZ A, et al. Index-based concatenated codes for the multi-draw DNA storage channel[C]. Proceedings of 2023 IEEE Information Theory Workshop. Saint-Malo, France, 2023: 383–388. doi: 10.1109/ITW55543.2023.10161631. [3] CAO Ben, ZHANG Xiaokang, CUI Shuang, et al. Adaptive coding for DNA storage with high storage density and low coverage[J]. npj Systems Biology and Applications, 2022, 8(1): 23. doi: 10.1038/s41540-022-00233-w. [4] MEISER L C, NGUYEN B H, CHEN Y J, et al. Synthetic DNA applications in information technology[J]. Nature Communications, 2022, 13(1): 352. doi: 10.1038/s41467-021-27846-9. [5] DORICCHI A, PLATNICH C M, GIMPEL A, et al. Emerging approaches to DNA data storage: Challenges and prospects[J]. ACS Nano, 2022, 16(11): 17552–17571. doi: 10.1021/acsnano.2c06748. [6] TOMEK K J, VOLKEL K, INDERMAUR E W, et al. Promiscuous molecules for smarter file operations in DNA-based data storage[J]. Nature Communications, 2021, 12(1): 3518. doi: 10.1038/s41467-021-23669-w. [7] EL-SHAIKH A, WELZEL M, HEIDER D, et al. High-scale random access on DNA storage systems[J]. NAR Genomics and Bioinformatics, 2022, 4(1): lqab126. doi: 10.1093/nargab/lqab126. [8] LUO Yuan, WANG Shuchen, FENG Zhuowei, et al. Integrated microfluidic DNA storage platform with automated sample handling and physical data partitioning[J]. Analytical Chemistry, 2022, 94(38): 13153–13162. doi: 10.1021/acs.analchem.2c02667. [9] TABATABAEI YAZDI S M, YUAN Yongbo, MA Jian, et al. A rewritable, random-access DNA-based storage system[J]. Scientific Reports, 2015, 5: 14138. doi: 10.1038/srep14138. [10] ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36(3): 242–248. doi: 10.1038/nbt.4079. [11] CHURCH G M, GAO Yuan, and KOSURI S. Next-generation digital information storage in DNA[J]. Science, 2012, 337(6102): 1628. doi: 10.1126/science.1226355. [12] BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system[C]. Proceedings of the Twenty-First International Conference on Architectural Support for Programming Languages and Operating Systems, New York, USA, 2016: 637–649. doi: 10.1145/2872362.2872397. [13] GRASS R N, HECKEL R, PUDDU M, et al. Robust chemical preservation of digital information on DNA in silica with error-correcting codes[J]. Angewandte Chemie International Edition, 2015, 54(8): 2552–2555. doi: 10.1002/anie.201411378. [14] BLAWAT M, GAEDKE K, HÜTTER I, et al. Forward error correction for DNA data storage[J]. Procedia Computer Science, 2016, 80: 1011–1022. doi: 10.1016/j.procs.2016.05.398. [15] ERLICH Y and ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950–954. doi: 10.1126/science.aaj2038. [16] GOLDMAN N, BERTONE P, CHEN Siyuan, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA[J]. Nature, 2013, 494(7435): 77–80. doi: 10.1038/nature11875. [17] SONG Xin, SHAH Shalin, and REIF J. Multidimensional data organization and random access in large-scale DNA storage systems[J]. Theoretical Computer Science, 2021, 894: 190–202. doi: 10.1016/j.tcs.2021.09.021. [18] NEWMAN S, STEPHENSON A P, WILLSEY M, et al. High density DNA data storage library via dehydration with digital microfluidic retrieval[J]. Nature Communications, 2019, 10(1): 1706. doi: 10.1038/s41467-019-09517-y. [19] CHOI Y, BAE H J, LEE A C, et al. DNA micro-disks for the management of DNA-based data storage with index and write-once–read-many (WORM) memory features[J]. Advanced Materials, 2020, 32(37): 2001249. doi: 10.1002/adma.202001249. [20] LAU B, CHANDAK S, ROY S, et al. Magnetic DNA random access memory with nanopore readouts and exponentially-scaled combinatorial addressing[J]. Scientific Reports, 2023, 13(1): 8514. doi: 10.1038/s41598-023-29575-z. [21] HEID C A, STEVENS J, LIVAK K J, et al. Real time quantitative PCR[J]. Genome Research, 1996, 6(10): 986–994. doi: 10.1101/gr.6.10.986. [22] BANCROFT C, BOWLER T, BLOOM B, et al. Long-term storage of information in DNA[J]. Science, 2001, 293(5536): 1763–1765. doi: 10.1126/science.293.5536.1763c. [23] SECILMIS L, TESTOLINA M, LAZZAROTTO D, et al. Towards effective visual information storage on DNA support[C]//Proceedings of SPIE 12226, Applications of Digital Image Processing XLV, San Diego, USA, 2022: 1222605. doi: 10.1117/12.2636678. -





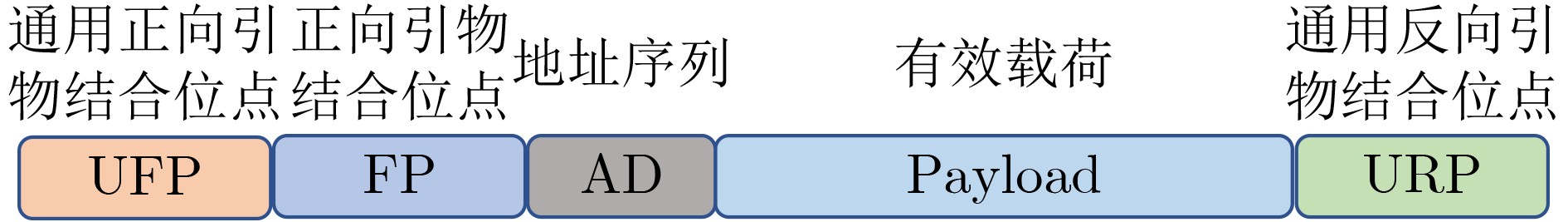
 下载:
下载:
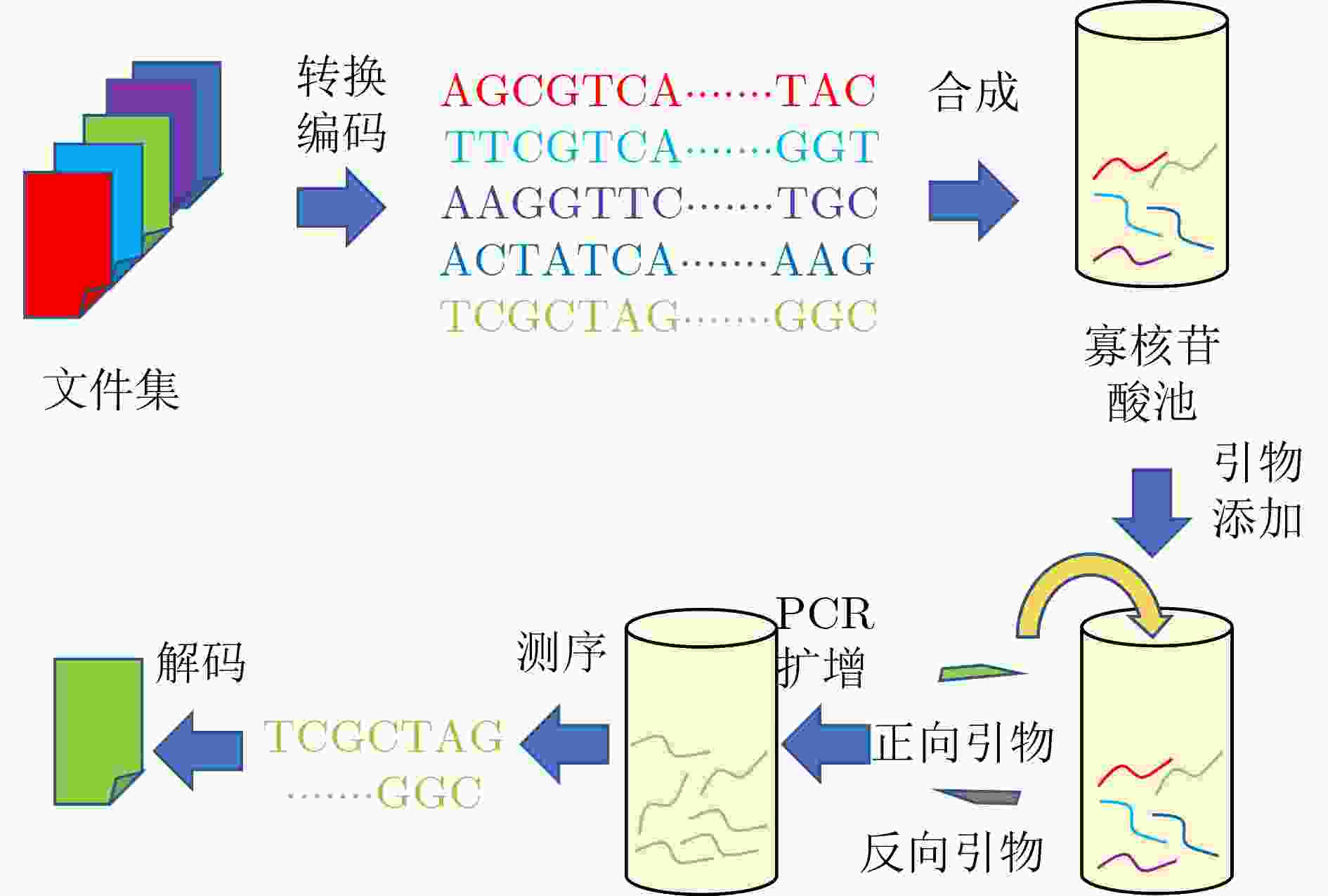
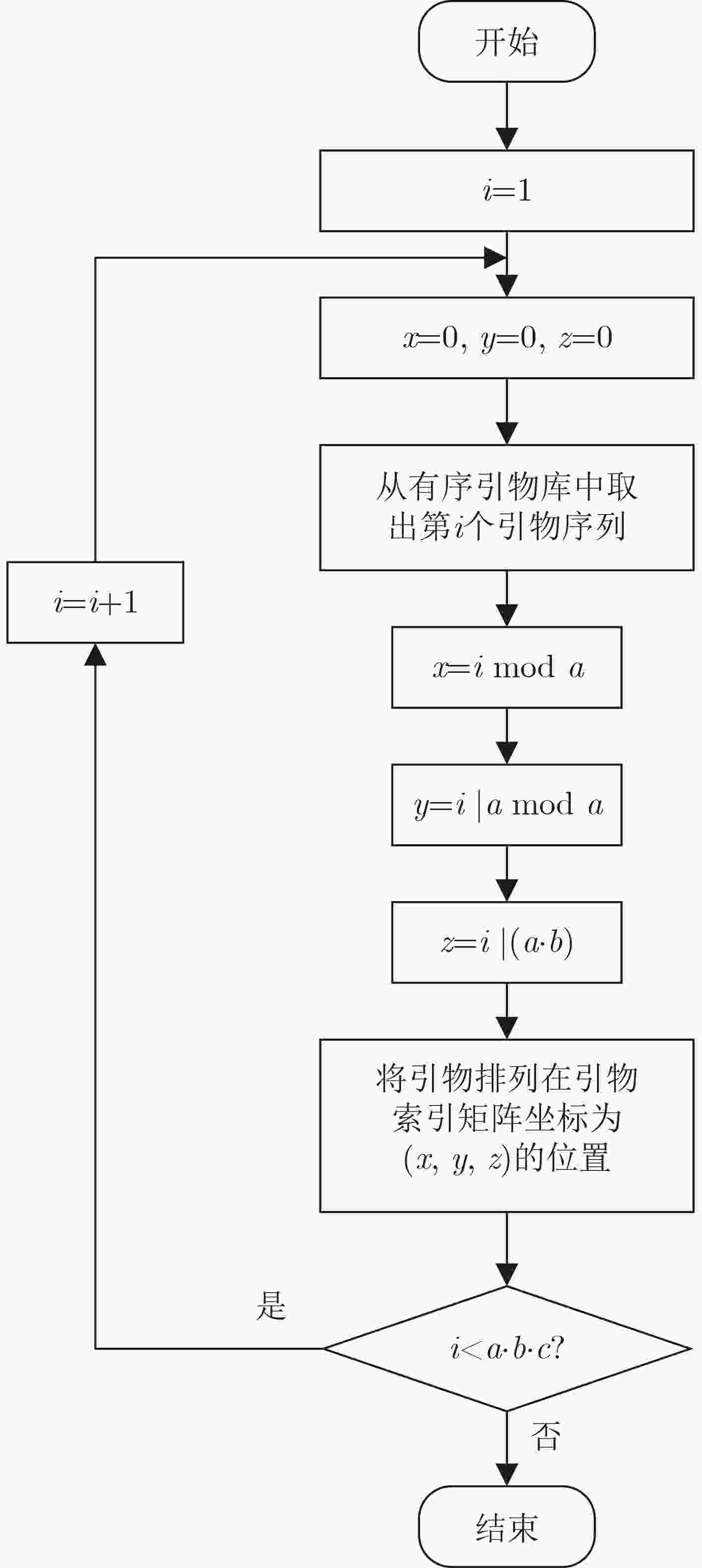
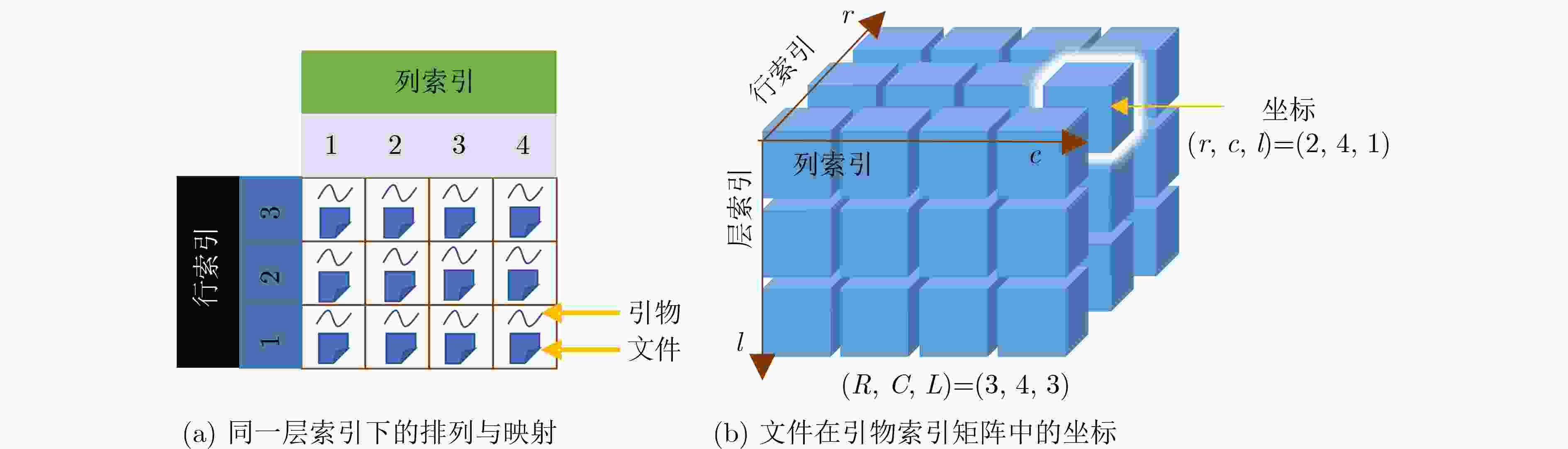

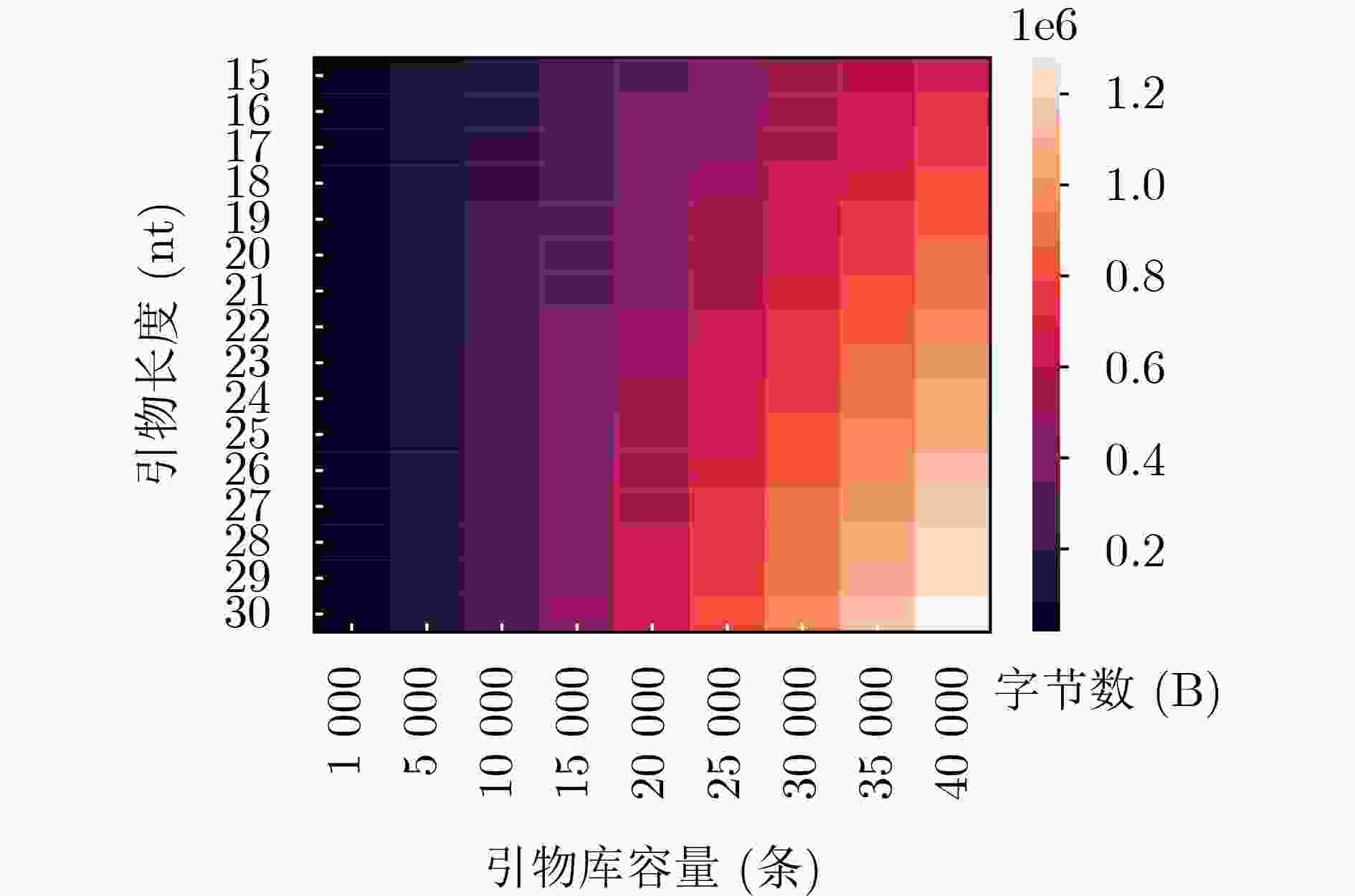
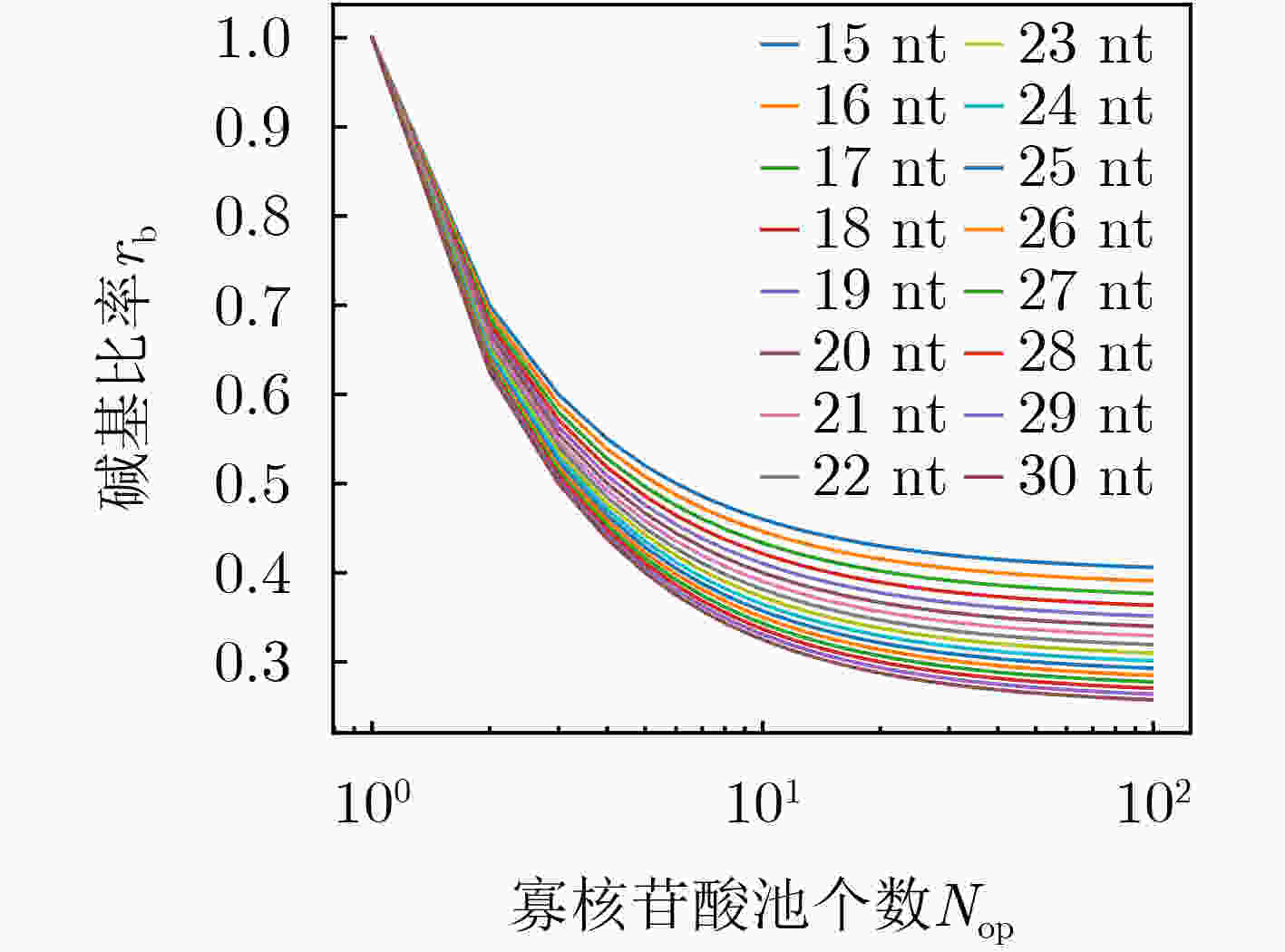
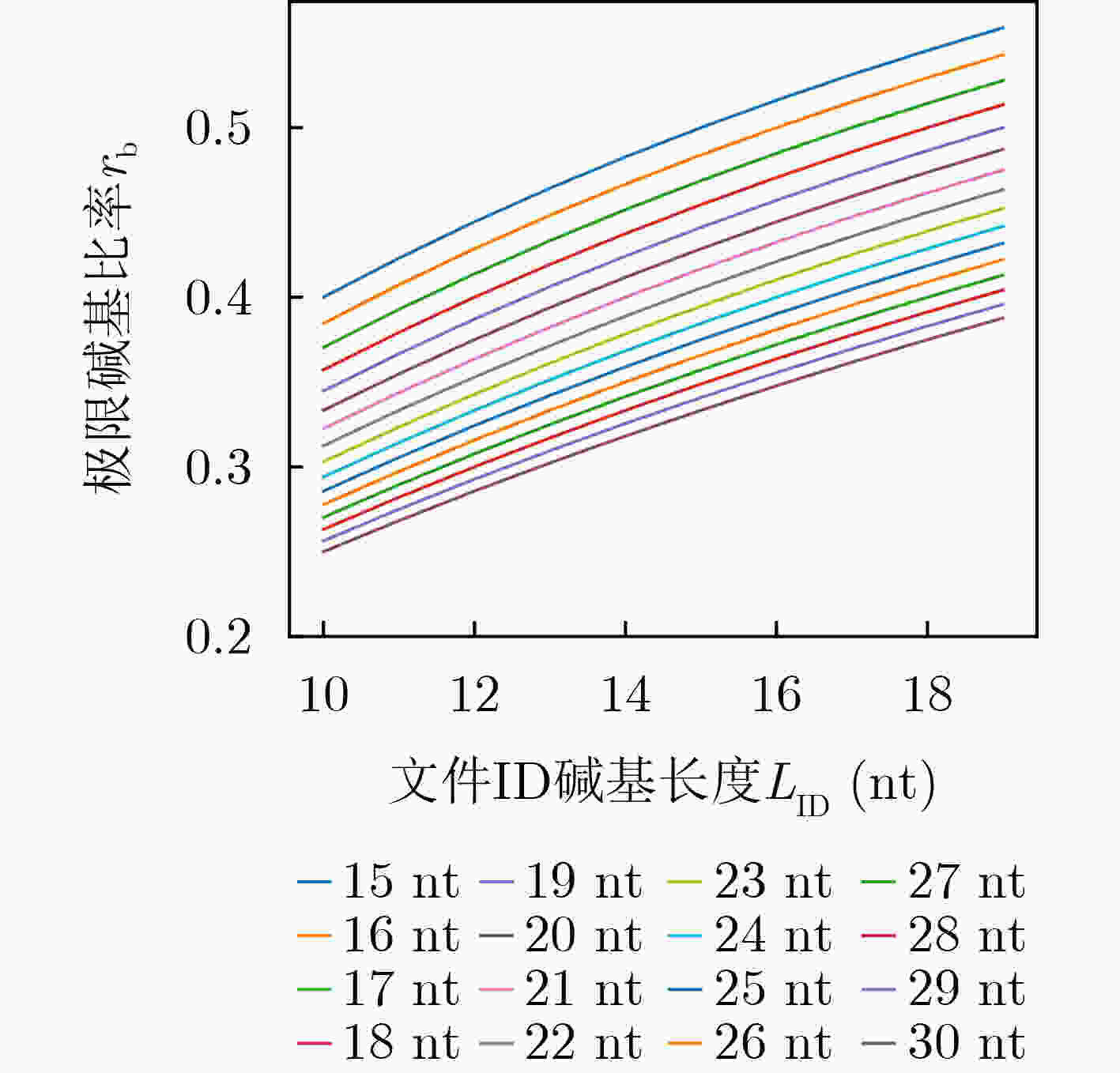
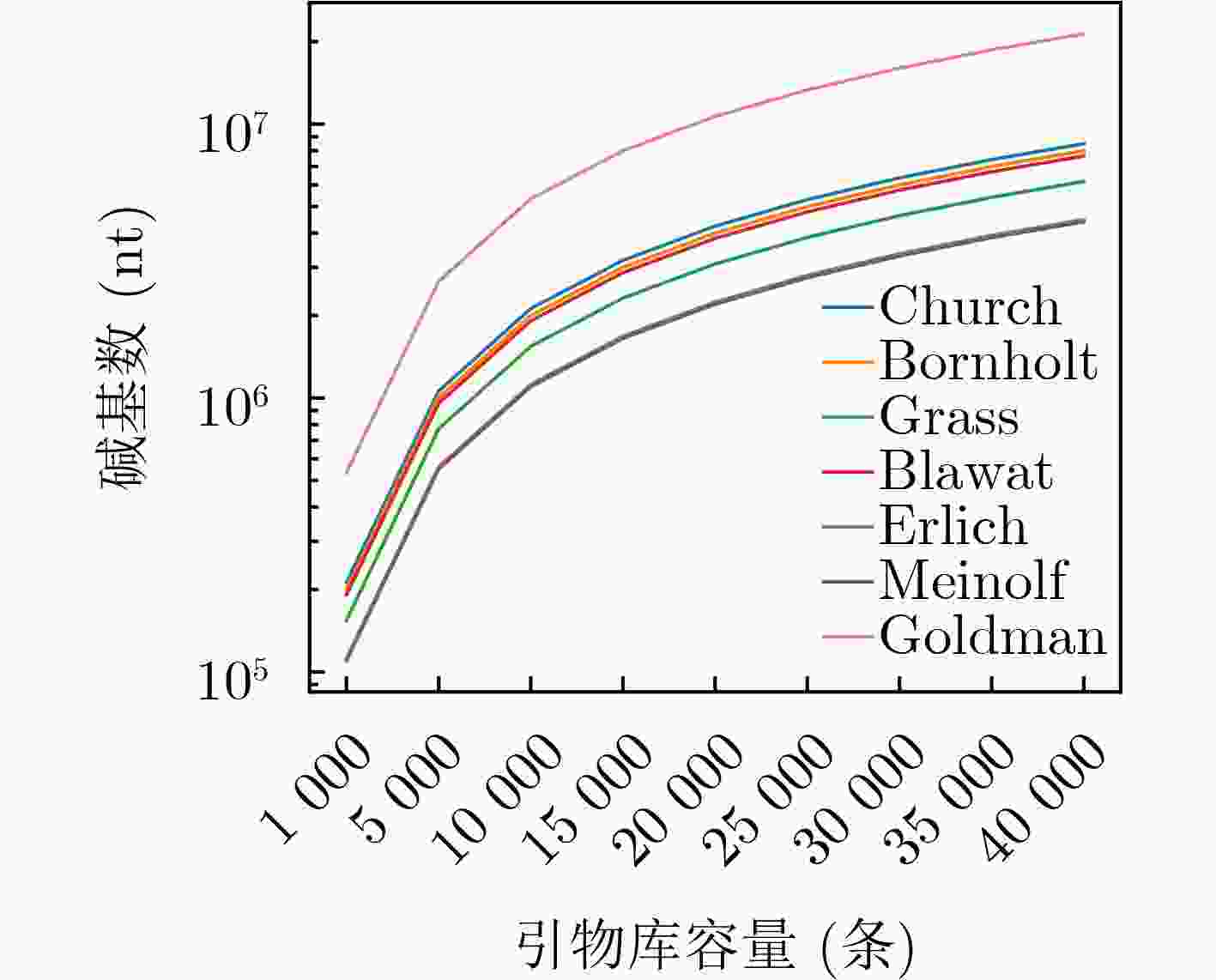


 下载:
下载:
