| [1] |
COLLINGER J L, WODLINGER B, DOWNEY J E, et al. High-performance neuroprosthetic control by an individual with tetraplegia[J]. The Lancet, 2013, 381(9866): 557–564. doi: 10.1016/S0140-6736(12)61816-9
|
| [2] |
SHANECHI M M, ORSBORN A L, MOORMAN H G, et al. Rapid control and feedback rates enhance neuroprosthetic control[J]. Nature Communications, 2017, 8: 13825. doi: 10.1038/ncomms13825
|
| [3] |
SEEBER B U and BRUCE I C. The history and future of neural modeling for cochlear implants[J]. Network: Computation in Neural Systems, 2016, 27(2/3): 53–66. doi: 10.1080/0954898X.2016.1223365
|
| [4] |
JOHNSON L A, DELLA SANTINA C C, and WANG Xiaoqin. Representations of time-varying cochlear implant stimulation in auditory cortex of awake marmosets (Callithrix jacchus)[J]. Journal of Neuroscience, 2017, 37(29): 7008–7022. doi: 10.1523/JNEUROSCI.0093-17.2017
|
| [5] |
GHEZZI D. Retinal prostheses: Progress toward the next generation implants[J]. Frontiers in Neuroscience, 2015, 9: 290. doi: 10.3389/fnins.2015.00290
|
| [6] |
TANG Jing, QIN Nan, CHONG Yan, et al. Nanowire arrays restore vision in blind mice[J]. Nature Communications, 2018, 9(1): 786. doi: 10.1038/s41467-018-03212-0
|
| [7] |
HUANG Tiejun, ZHENG Yajing, YU Zhaofei, et al. 1000× faster camera and machine vision with ordinary devices[J]. Engineering, To be published.
|
| [8] |
ZHU Lin, DONG Siwei, LI Jianing, et al. Retina-like visual image reconstruction via spiking neural model[C]. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, USA, 2020: 1438–1446.
|
| [9] |
ZHENG Yajing, ZHENG Lingxiao, YU Zhaofei, et al. High-speed image reconstruction through short-term plasticity for spiking cameras[C]. 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, USA, 2021: 6354–6363.
|
| [10] |
ZHAO Jing, XIONG Ruiqin, XIE Jiyu, et al. Reconstructing clear image for high-speed motion scene with a retina-inspired spike camera[J]. IEEE Transactions on Computational Imaging, 2022, 8: 12–27. doi: 10.1109/TCI.2021.3136446
|
| [11] |
ZHAO Junwei, YU Zhaofei, MA Lei, et al. Modeling the detection capability of high-speed spiking cameras[C]. 2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 2022: 4653–4657.
|
| [12] |
DING Ziluo, ZHAO Rui, ZHANG Jiyuan, et al. Spatio-temporal recurrent networks for event-based optical flow estimation[C]. The 36th AAAI Conference on Artificial Intelligence, Palo Alto, USA, 2022: 525–533.
|
| [13] |
HU Liwen, ZHAO Rui, DING Ziluo, et al. Optical flow estimation for spiking camera[C]. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, USA, 2022: 17844–17853.
|
| [14] |
LINDEN J F, LIU R C, SAHANI M, et al. Spectrotemporal structure of receptive fields in areas AI and AAF of mouse auditory cortex[J]. Journal of Neurophysiology, 2003, 90(4): 2660–2675. doi: 10.1152/jn.00751.2002
|
| [15] |
MACHENS C K, WEHR M S, and ZADOR A M. Linearity of cortical receptive fields measured with natural sounds[J]. Journal of Neuroscience, 2004, 24(5): 1089–1100. doi: 10.1523/JNEUROSCI.4445-03.2004
|
| [16] |
SAHANI M and LINDEN J F. How linear are auditory cortical responses?[C]. The 15th International Conference on Neural Information Processing Systems, Vancouver, Canada, 2002: 125–132.
|
| [17] |
SHARPEE T, RUST N C, and BIALEK W. Analyzing neural responses to natural signals: Maximally informative dimensions[J]. Neural Computation, 2004, 16(2): 223–250. doi: 10.1162/089976604322742010
|
| [18] |
CHICHILNISKY E J. A simple white noise analysis of neuronal light responses[J]. Network, 2001, 12(2): 199–213. doi: 10.1080/713663221
|
| [19] |
PANINSKI L. Maximum likelihood estimation of cascade point-process neural encoding models[J]. Network: Computation in Neural Systems, 2004, 15(4): 243–262. doi: 10.1088/0954-898X_15_4_002
|
| [20] |
RABINOWITZ N C, WILLMORE B D B, SCHNUPP J W H, et al. Contrast gain control in auditory cortex[J]. Neuron, 2011, 70(6): 1178–1191. doi: 10.1016/j.neuron.2011.04.030
|
| [21] |
VINJE W E and GALLANT J L. Natural stimulation of the nonclassical receptive field increases information transmission efficiency in V1[J]. Journal of Neuroscience, 2002, 22(7): 2904–2915. doi: 10.1523/JNEUROSCI.22-07-02904.2002
|
| [22] |
LIU J K, SCHREYER H M, ONKEN A, et al. Inference of neuronal functional circuitry with spike-triggered non-negative matrix factorization[J]. Nature Communications, 2017, 8(1): 149. doi: 10.1038/s41467-017-00156-9
|
| [23] |
SHAH N P, BRACKBILL N, RHOADES C, et al. Inference of nonlinear receptive field subunits with spike-triggered clustering[J]. eLife, 2020, 9: e45743. doi: 10.7554/eLife.45743
|
| [24] |
KARAMANLIS D and GOLLISCH T. Nonlinear spatial integration underlies the diversity of retinal ganglion cell responses to natural images[J]. Journal of Neuroscience, 2021, 41(15): 3479–3498. doi: 10.1523/JNEUROSCI.3075-20.2021
|
| [25] |
LIU J K, KARAMANLIS D, and GOLLISCH T. Simple model for encoding natural images by retinal ganglion cells with nonlinear spatial integration[J]. PLoS Computational Biology, 2022, 18(3): e1009925. doi: 10.1371/journal.pcbi.1009925
|
| [26] |
MCFARLAND J M, CUI Yuwei, and BUTTS D A. Inferring nonlinear neuronal computation based on physiologically plausible inputs[J]. PLoS Computational Biology, 2013, 9(7): e1003143. doi: 10.1371/journal.pcbi.1003143
|
| [27] |
DORRN A L, YUAN Kexin, BARKER A J, et al. Developmental sensory experience balances cortical excitation and inhibition[J]. Nature, 2010, 465(7300): 932–936. doi: 10.1038/nature09119
|
| [28] |
MARMARELIS V. Analysis of Physiological Systems: The White-Noise Approach[M]. Springer, 2012.
|
| [29] |
PARK I M, ARCHER E, PRIEBE N, et al. Spectral methods for neural characterization using generalized quadratic models[C]. The 26th International Conference on Neural Information Processing Systems, Lake Tahoe USA, 2013: 2454–2462.
|
| [30] |
PARK I M and PILLOW J W. Bayesian spike-triggered covariance analysis[C]. The 24th International Conference on Neural Information Processing Systems, Granada, Spain, 2011: 1692–1700.
|
| [31] |
JIA Shanshan, XING Dajun, YU Zhaofei, et al. Dissecting cascade computational components in spiking neural networks[J]. PLoS Computational Biology, 2021, 17(11): e1009640. doi: 10.1371/journal.pcbi.1009640
|
| [32] |
LECUN Y, BENGIO Y, and HINTON G. Deep learning[J]. Nature, 2015, 521(7553): 436–444. doi: 10.1038/nature14539
|
| [33] |
YAMINS D L K, HONG Ha, CADIEU C F, et al. Performance-optimized hierarchical models predict neural responses in higher visual cortex[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(23): 8619–8624. doi: 10.1073/pnas.1403112111
|
| [34] |
KHALIGH-RAZAVI S M and KRIEGESKORTE N. Deep supervised, but not unsupervised, models may explain IT cortical representation[J]. PLoS Computational Biology, 2014, 10(11): e1003915. doi: 10.1371/journal.pcbi.1003915
|
| [35] |
KRIEGESKORTE N. Deep neural networks: A new framework for modeling biological vision and brain information processing[J]. Annual Review of Vision Science, 2015, 1: 417–446. doi: 10.1146/annurev-vision-082114-035447
|
| [36] |
YAMINS D L K and DICARLO J J. Using goal-driven deep learning models to understand sensory cortex[J]. Nature Neuroscience, 2016, 19(3): 356–365. doi: 10.1038/nn.4244
|
| [37] |
ROWEKAMP R J and SHARPEE T O. Cross-orientation suppression in visual area V2[J]. Nature Communications, 2017, 8: 15739. doi: 10.1038/ncomms15739
|
| [38] |
CADENA S A, DENFIELD G H, WALKER E Y, et al. Deep convolutional models improve predictions of macaque V1 responses to natural images[J]. PLoS Computational Biology, 2019, 15(4): e1006897. doi: 10.1371/journal.pcbi.1006897
|
| [39] |
YAN Qi, ZHENG Yajing, JIA Shanshan, et al. Revealing fine structures of the retinal receptive field by deep-learning networks[J]. IEEE Transactions on Cybernetics, 2022, 52(1): 39–50. doi: 10.1109/TCYB.2020.2972983
|
| [40] |
VANCE P J, DAS G P, KERR D, et al. Bioinspired approach to modeling retinal ganglion cells using system identification techniques[J]. IEEE Transactions on Neural Networks and Learning Systems, 2018, 29(5): 1796–1808. doi: 10.1109/TNNLS.2017.2690139
|
| [41] |
MCINTOSH L T, MAHESWARANATHAN N, NAYEBI A, et al. Deep learning models of the retinal response to natural scenes[C]. The 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 2016: 1369–1377.
|
| [42] |
KAR K, KUBILIUS J, SCHMIDT K, et al. Evidence that recurrent circuits are critical to the ventral stream’ s execution of core object recognition behavior[J]. Nature Neuroscience, 2019, 22(6): 974–983. doi: 10.1038/s41593-019-0392-5
|
| [43] |
KIETZMANN T C, SPOERER C J, SÖRENSEN L K A, et al. Recurrence is required to capture the representational dynamics of the human visual system[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(43): 21854–21863. doi: 10.1073/pnas.1905544116
|
| [44] |
RAJAEI K, MOHSENZADEH Y, EBRAHIMPOUR R, et al. Beyond core object recognition: Recurrent processes account for object recognition under occlusion[J]. PLoS Computational Biology, 2019, 15(5): e1007001. doi: 10.1371/journal.pcbi.1007001
|
| [45] |
LINSLEY D, KIM J, VEERABADRAN V, et al. Learning long-range spatial dependencies with horizontal gated recurrent units[C]. The 32nd International Conference on Neural Information Processing Systems, Montréal, Canada, 2018: 152–164.
|
| [46] |
O’BRIEN J and BLOOMFIELD S A. Plasticity of retinal gap junctions: Roles in synaptic physiology and disease[J]. Annual Review of Vision Science, 2018, 4: 79–100. doi: 10.1146/annurev-vision-091517-034133
|
| [47] |
RIVLIN-ETZION M, GRIMES W N, and RIEKE F. Flexible neural hardware supports dynamic computations in retina[J]. Trends in Neurosciences, 2018, 41(4): 224–237. doi: 10.1016/j.tins.2018.01.009
|
| [48] |
TRENHOLM S, SCHWAB D J, BALASUBRAMANIAN V, et al. Lag normalization in an electrically coupled neural network[J]. Nature Neuroscience, 2013, 16(2): 154–156. doi: 10.1038/nn.3308
|
| [49] |
YU Zhaofei, LIU J K, JIA Shanshan, et al. Toward the next generation of retinal neuroprosthesis: Visual computation with spikes[J]. Engineering, 2020, 6(4): 449–461. doi: 10.1016/j.eng.2020.02.004
|
| [50] |
ZHENG Yajing, JIA Shanshan, YU Zhaofei, et al. Unraveling neural coding of dynamic natural visual scenes via convolutional recurrent neural networks[J]. Patterns, 2021, 2(10): 100350. doi: 10.1016/j.patter.2021.100350
|
| [51] |
PANINSKI L. Convergence properties of some spike-triggered analysis techniques[C]. The 15th International Conference on Neural Information Processing Systems, Vancouver, British Columbia, Canada, 2002: 189–196.
|
| [52] |
JIA Shanshan, YU Zhaofei, ONKEN A, et al. Neural system identification with spike-triggered non-negative matrix factorization[J]. IEEE Transactions on Cybernetics, 2022, 52(6): 4772–4783. doi: 10.1109/TCYB.2020.3042513
|
| [53] |
ONKEN A, LIU J K, KARUNASEKARA P P C R, et al. Using matrix and tensor factorizations for the single-trial analysis of population spike trains[J]. PLoS Computational Biology, 2016, 12(11): e1005189. doi: 10.1371/journal.pcbi.1005189
|
| [54] |
WILLIAMS A H, KIM T H, WANG F, et al. Unsupervised discovery of demixed, low-dimensional neural dynamics across multiple timescales through tensor component analysis[J]. Neuron, 2018, 98(6): 1099–1115.e8. doi: 10.1016/j.neuron.2018.05.015
|
| [55] |
ZHUANG Chengxu, YAN Siming, NAYEBI A, et al. Unsupervised neural network models of the ventral visual stream[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(3): e2014196118. doi: 10.1073/pnas.2014196118
|
| [56] |
BRENNER N, STRONG S P, KOBERLE R, et al. Synergy in a neural code[J]. Neural Computation, 2000, 12(7): 1531–1552. doi: 10.1162/089976600300015259
|
| [57] |
SHARPEE T O, MILLER K D, and STRYKER M P. On the importance of static nonlinearity in estimating spatiotemporal neural filters with natural stimuli[J]. Journal of Neurophysiology, 2008, 99(5): 2496–2509. doi: 10.1152/jn.01397.2007
|
| [58] |
MEYER A F, DIEPENBROCK J P, HAPPEL M F K, et al. Discriminative learning of receptive fields from responses to non-Gaussian stimulus ensembles[J]. PLoS One, 2014, 9(4): e93062. doi: 10.1371/journal.pone.0093062
|
| [59] |
MEYER A F, DIEPENBROCK J P, OHL F W, et al. Quantifying neural coding noise in linear threshold models[C]. The 6th International IEEE/EMBS Conference on Neural Engineering, San Diego, USA, 2013: 1127–1130.
|
| [60] |
ZAPP S J, NITSCHE S, and GOLLISCH T. Retinal receptive-field substructure: Scaffolding for coding and computation[J]. Trends in Neurosciences, 2022, 45(6): 430–445. doi: 10.1016/J.TINS.2022.03.005
|
| [61] |
KARAMANLIS D, SCHREYER H M, and GOLLISCH T. Retinal encoding of natural scenes[J]. Annual Review of Vision Science, 2022, 8: 171–193. doi: 10.1146/annurev-vision-100820-114239
|
| [62] |
SALAHIAN N, TAB F A, SEYEDI S A, et al. Deep autoencoder-like NMF with contrastive regularization and feature relationship preservation[J]. Expert Systems with Applications, 2023, 214: 119051. doi: 10.1016/J.ESWA.2022.119051
|
| [63] |
CHEN Wensheng, ZENG Qianwen, and PAN Binbin. A survey of deep nonnegative matrix factorization[J]. Neurocomputing, 2022, 491: 305–320. doi: 10.1016/j.neucom.2021.08.152
|
| [64] |
XU Qi, LI Yaxin, SHEN Jiangrong, et al. Hierarchical spiking-based model for efficient image classification with enhanced feature extraction and encoding[J]. IEEE Transactions on Neural Networks and Learning Systems, To be published.
|





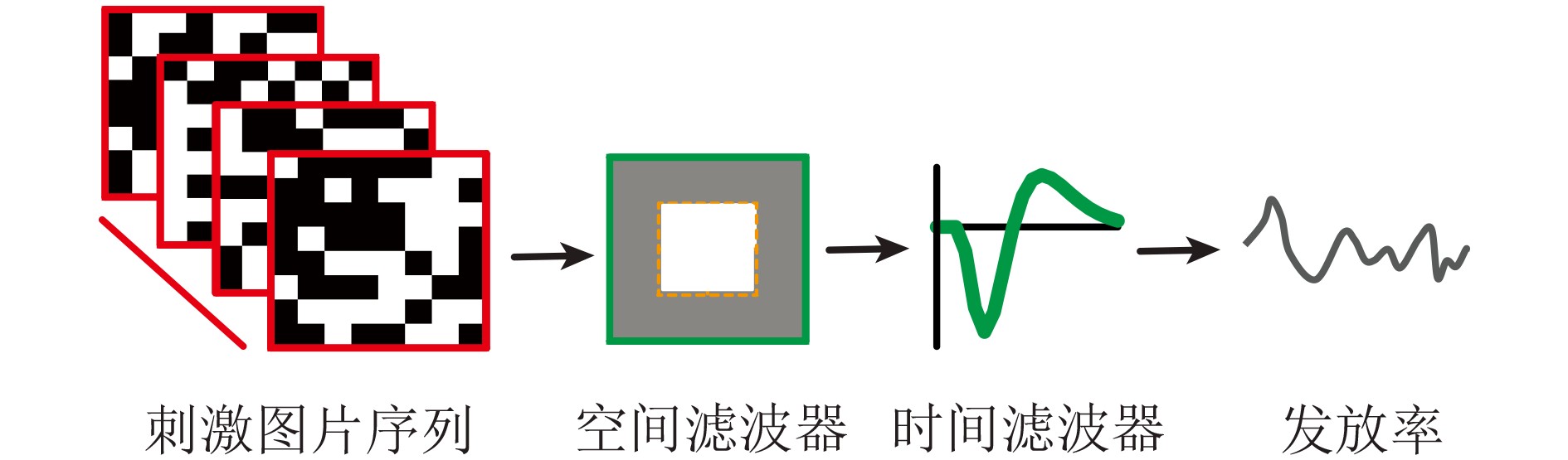
 下载:
下载:
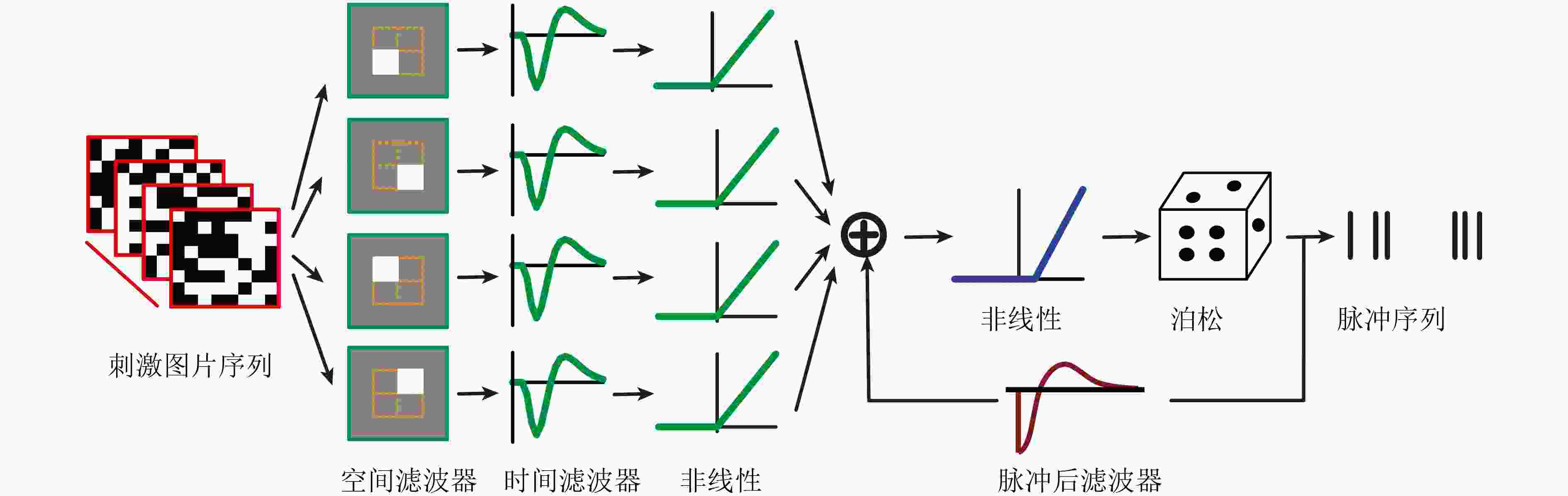


 下载:
下载:
