A Transformer Segmentation Model for PET/CT Images with Cross-modal, Cross-scale and Cross-dimensional
-
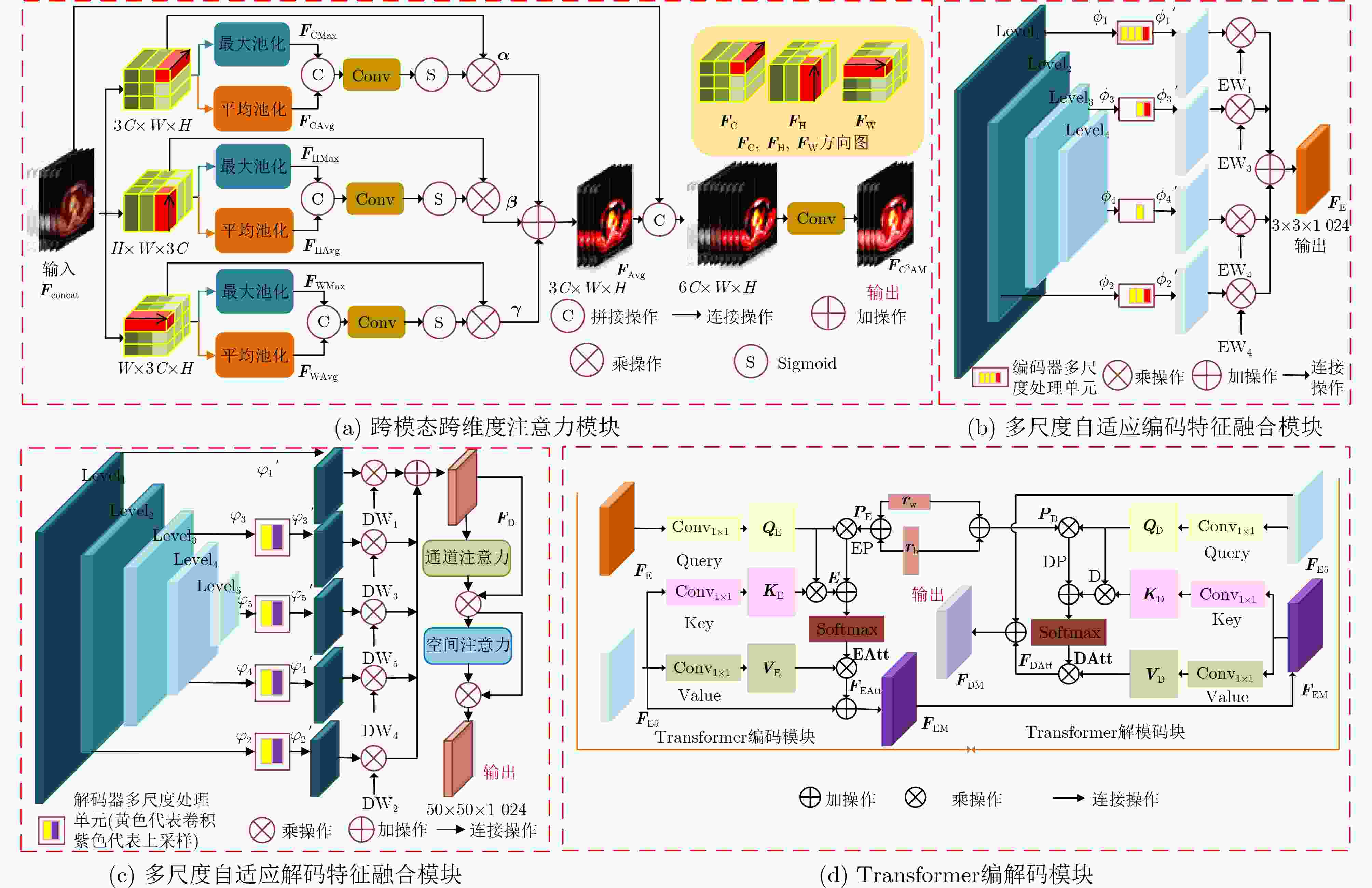
摘要: 多模态医学图像能够有效融合解剖图像和功能图像的信息,将人体内部的功能、解剖等多方面信息反映在同一幅图像上,在临床上有十分重要的意义。针对如何高效利用多模态医学图像信息的综合表达能力,以及如何充分提取跨尺度上下文信息的问题,该文提出跨模态跨尺度跨维度的PET/CT图像的Transformer分割模型。该模型主要改进是,首先,在编码器部分设计了PET/CT主干分支和PET, CT辅助分支提取多模态图像信息;然后,在跳跃连接部分设计了跨模态跨维度注意力模块从模态和维度角度出发捕获跨模态图像各维的有效信息;其次,在瓶颈层构造跨尺度Transformer模块,自适应融合深层的语义信息和浅层的空间信息使网络学习到更多的上下文信息,并从中获取跨尺度全局信息;最后,在解码器部分提出多尺度自适应解码特征融合模块,聚合并充分利用解码路径得到精细程度不同的多尺度特征图,缓解上采样引入的噪声。在临床多模态肺部医学图像数据集验证算法的有效性,结果表明所提模型对于肺部病灶分割的Acc, Recall, Dice, Voe, Rvd和Miou分别为97.99%, 94.29%, 95.32%, 92.74%, 92.95%和90.14%,模型对于形状复杂的病灶分割具有较高的精度和相对较低的冗余度。
-
关键词:
- 医学图像分割 /
- 跨模态 /
- 跨维度 /
- Transformer /
- U型网络
Abstract: Multi-modal medical images can effectively fuse anatomical images and functional images. It reflects the functional and anatomical information within the body on the same image, which gives rise to critical clinical implications. How to utilize efficiently the comprehensive representation capabilities of multimodal medical image information and how to extract adequately cross-scale contextual information are key questions. In this paper, a Transformer segmentation model for PET/CT images with cross-modal, cross-scale and cross-dimension is proposed. The main improvements of the model are as follows: Firstly, PET/CT backbone branch, PET auxiliary branch, and CT auxiliary branch are designed to extract multi-modal image information in the encoder section; Secondly, a cross-modal and cross-dimensional attention module is designed in the skip connection part. The valid information in each dimension of the cross-modal images is captured by this module from both modal and dimensional views; Thirdly, a cross-scale Transformer module is designed at the bottleneck level. Deep semantic information and shallow spatial information are adaptively fused by this model, which can enable the network to learn more contextual information and obtain cross-scale global information; Finally, a multi-scale adaptive decoding feature fusion module is proposed in the decoder part. The multi-scale feature maps with different levels of detail are aggregated and fully utilized in the decoding path, and the noise introduced by upsampling is mitigated in this module. The effectiveness of the algorithm is verified by using a clinical multi-modal lung medical image dataset. All results show that the Acc, Recall, Dice, Voe, Rvd and Miou of the proposed model for lung lesion segmentation are: 97.99%, 94.29%, 95.32%, 92.74%, 92.95% and 90.14%. For the segmentation of lung lesions with complex shapes, it has high accuracy and relatively low redundancy.-
Key words:
- Medical image segmentation /
- Cross-modal /
- Cross-dimensional /
- Transformer /
- U-Net
-
表 1 评价指标
评价指标 定义 评价指标 定义 Acc ${\rm{Acc}} = \dfrac{ \rm{TP + FN} }{ \rm{TP + TN + FP + FN} }$ Voe ${\rm{Voe} } = {\rm{abs} }\left( {1 - \left| {\dfrac{ { {{P} } \cap {{G} } } }{ { {{P} } \cup {{G} } } } } \right|} \right)$ Recall ${\rm{Recall}} = \dfrac{ \rm{TP} }{ \rm{TP + FN} }$ Rvd ${\rm{Rvd} } = {\rm{abs} }\left( {\dfrac{ {\left( { {{P} } - {{G} } } \right)} }{ {{G} } } } \right)$ Dice ${\rm{Dice}} = \dfrac{ {2 \times {\rm{TP}}} }{ \rm{FN + TP + TP + FP} }$ Miou ${\rm{Miou}} = \dfrac{ {{\rm{TP}}} }{ {{\rm{FN}} + {\rm{TP}} + {\rm{FP}}} }$ 表 2 跨模态相关性实验分割结果(%)
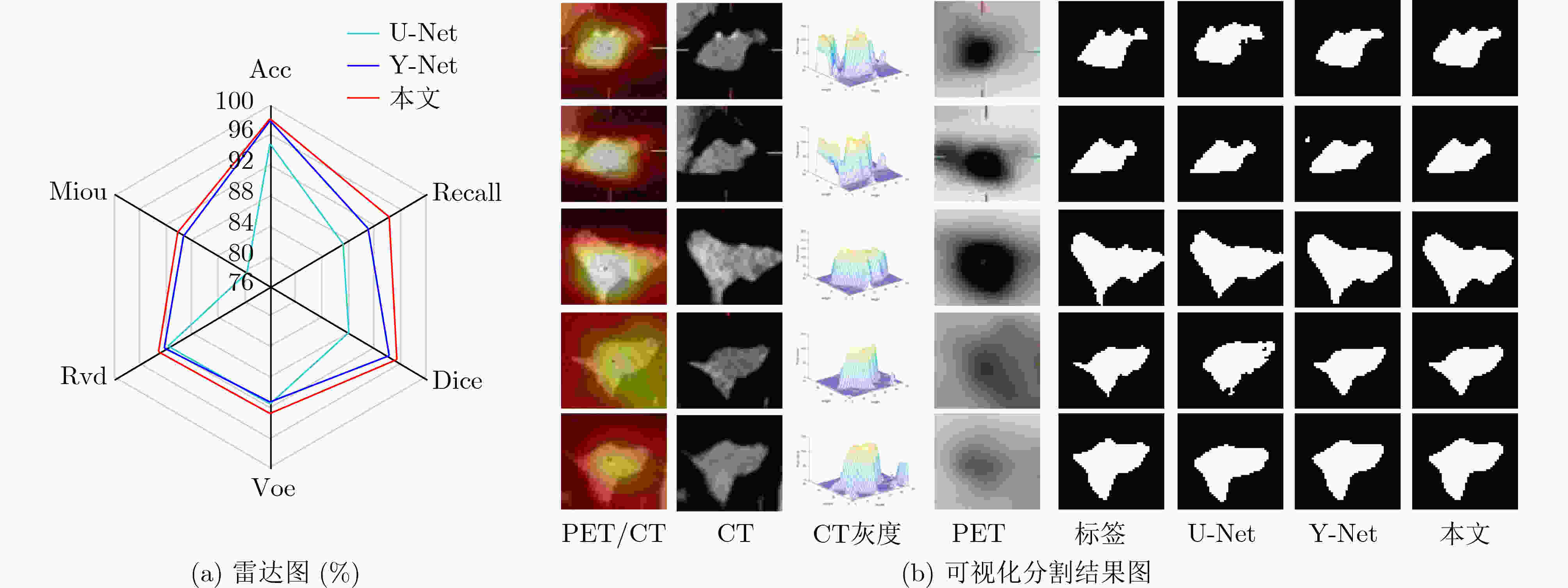
表 3 不同分割网络对比实验分割结果(%)
模型 Acc Recall Dice Voe Rvd Miou UNet++[13] 92.19 80.67 82.03 85.61 86.23 70.62 SegNet[14] 92.09 81.96 82.25 86.88 86.90 70.60 Attention UNet [15] 92.37 80.91 82.30 86.03 86.68 71.11 SEResUNet[16] 92.78 81.56 83.45 88.15 88.58 72.60 MsTGANet[10] 97.29 81.48 83.34 86.56 87.28 72.54 MEAU-Net[3] 96.84 93.76 92.76 88.35 86.93 86.89 本文 97.99 94.29 95.32 92.74 92.95 90.14 表 4 跨模跨尺度跨维度实验分割结果(%)
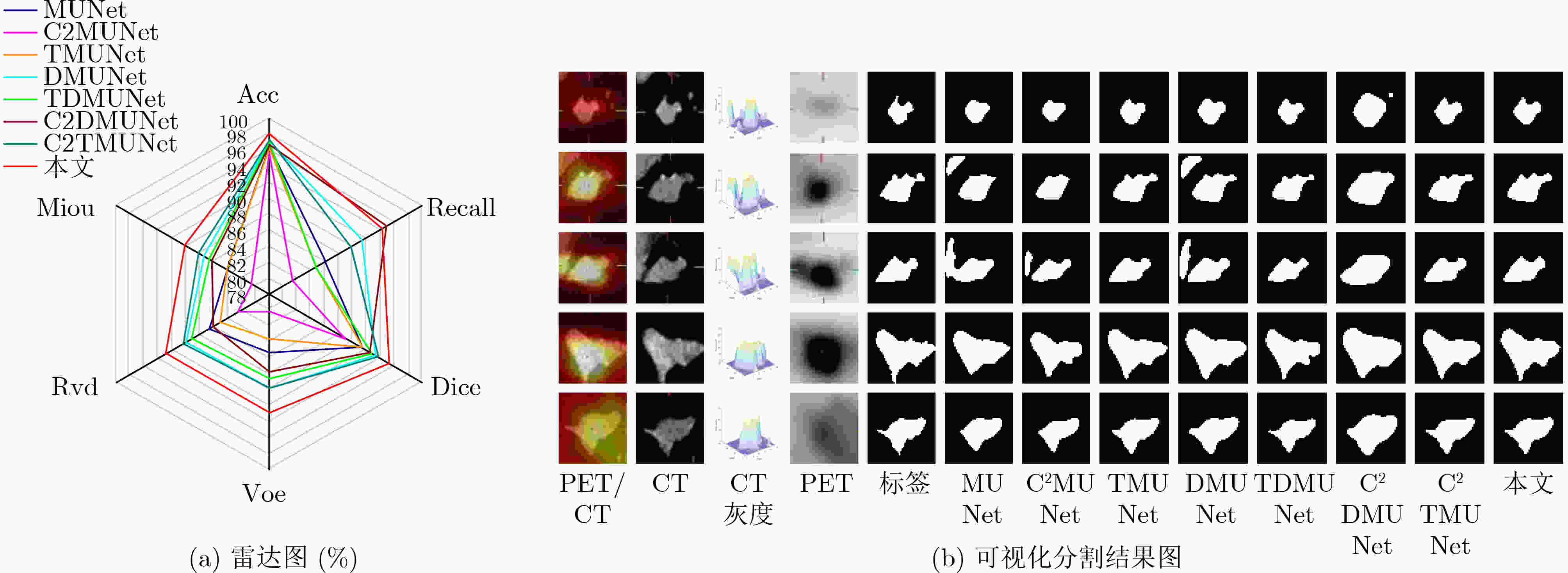
模型 Acc Recall Dice Voe Rvd Miou MUNet 96.44 85.87 91.18 85.34 86.56 84.15 C2MUNet 95.61 81.44 88.95 80.15 82.38 80.60 TMUNet 96.35 84.65 91.15 83.52 85.12 84.03 DMUNet 97.18 91.43 93.22 89.73 89.99 87.52 TDMUNet 96.97 84.65 92.70 88.51 89.17 86.62 C2DMUNet 96.57 94.78 92.40 87.60 85.90 86.34 C2TMUNet 97.33 89.71 93.57 89.66 90.40 88.11 本文 97.99 94.29 95.32 92.74 92.95 90.14 -
[1] WANG Xue, LI Zhanshan, HUANG Yongping, et al. Multimodal medical image segmentation using multi-scale context-aware network[J]. Neurocomputing, 2022, 486: 135–146. doi: 10.1016/j.neucom.2021.11.017 [2] FU Xiaohang, LEI Bi, KUMAR A, et al. Multimodal spatial attention module for targeting multimodal PET-CT lung tumor segmentation[J]. IEEE Journal of Biomedical and Health Informatics, 2021, 25(9): 3507–3516. doi: 10.1109/JBHI.2021.3059453 [3] 周涛, 董雅丽, 刘珊, 等. 用于肺部肿瘤图像分割的跨模态多编码混合注意力U-Net[J]. 光子学报, 2022, 51(4): 0410006. doi: 10.3788/gzxb20225104.0410006ZHOU Tao, DONG Yali, LIU Shan, et al. Cross-modality multi-encoder hybrid attention U-Net for lung tumors images segmentation[J]. Acta Photonica Sinica, 2022, 51(4): 0410006. doi: 10.3788/gzxb20225104.0410006 [4] KUMAR A, FULHAM M, FENG Dagan, et al. Co-learning feature fusion maps from PET-CT images of lung cancer[J]. IEEE Transactions on Medical Imaging, 2020, 39(1): 204–217. doi: 10.1109/TMI.2019.2923601 [5] 田永林, 王雨桐, 王建功, 等. 视觉Transformer研究的关键问题: 现状及展望[J]. 自动化学报, 2022, 48(4): 957–979. doi: 10.16383/j.aas.c220027TIAN Yonglin, WANG Yutong, WANG Jiangong, et al. Key problems and progress of vision transformers: The state of the art and prospects[J]. Acta Automatica Sinica, 2022, 48(4): 957–979. doi: 10.16383/j.aas.c220027 [6] 胡敏, 周秀东, 黄宏程, 等. 基于改进U型神经网络的脑出血CT图像分割[J]. 电子与信息学报, 2022, 44(1): 127–137. doi: 10.11999/JEIT200996HU Min, ZHOU Xiudong, HUANG Hongcheng, et al. Computed-tomography image segmentation of cerebral hemorrhage based on improved U-shaped neural network[J]. Journal of Electronics &Information Technology, 2022, 44(1): 127–137. doi: 10.11999/JEIT200996 [7] WANG Ziyue, PENG Yanjun, LI Dapeng, et al. MMNet: A multi-scale deep learning network for the left ventricular segmentation of cardiac MRI images[J]. Applied Intelligence, 2022, 52(5): 5225–5240. doi: 10.1007/s10489-021-02720-9 [8] ZHOU Tao, YE Xinyu, LU Huiling, et al. Dense convolutional network and its application in medical image analysis[J]. BioMed Research International, 2022, 2022: 2384830. doi: 10.1155/2022/2384830 [9] LIU Songtao, HUANG Di, and WANG Yunhong. Learning spatial fusion for single-shot object detection[EB/OL]. https://arxiv.org/abs/1911.09516v1, 2019. [10] WANG Meng, ZHU Weifang, SHI Fei, et al. MsTGANet: Automatic drusen segmentation from retinal OCT images[J]. IEEE Transactions on Medical Imaging, 2022, 41(2): 394–406. doi: 10.1109/TMI.2021.3112716 [11] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. The 18th International Conference on Medical Image Computing and Computer-assisted Intervention, Munich, Germany, 2015: 234–241. [12] LAN Hengrong, JIANG Daohuai, YANG Changchun, et al. Y-Net: Hybrid deep learning image reconstruction for photoacoustic tomography in vivo[J]. Photoacoustics, 2020, 20: 100197. doi: 10.1016/j.pacs.2020.100197 [13] ZHOU Zongwei, SIDDIQUEE M R, TAJBAKHSH N, et al. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation[J]. IEEE Transactions on Medical Imaging, 2020, 39(6): 1856–1867. doi: 10.1109/TMI.2019.2959609 [14] BADRINARAYANAN V, KENDALL A, and CIPOLLA R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation[J]. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2017, 39(12): 2481–2495. doi: 10.1109/TPAMI.2016.2644615 [15] OKTAY O, SCHLEMPER J, LE FOLGOC L, et al. Attention U-Net: Learning where to look for the pancreas[EB/OL]. https://arxiv.org/abs/1804.03999, 2018. [16] CAO Zheng, YU Bohan, LEI Biwen, et al. Cascaded SE-ResUnet for segmentation of thoracic organs at risk[J]. Neurocomputing, 2021, 453: 357–368. doi: 10.1016/j.neucom.2020.08.086 -






 下载:
下载:



 下载:
下载:
