An Efficient Bueket-allocation Decoding Method Based on Forward Error Correction Codes for Deoxyribo Nucleicecid Storage
-
摘要: 与传统存储方式相比,脱氧核糖核酸(DNA)存储的难点是测序序列中的插入和删除错误给信息解码过程带来了巨大挑战。针对具有1位纠错能力的前向纠错编码DNA存储,该文提出一种桶式分配策略提高解码的精度和效率。首先,搜索每个分组中所有测序读长的可识别DNA码,根据1位纠错能力确定其对应的合法编码;其次,根据每个可识别DNA码在测序读长的位置确定相应编码的最佳编码位置(即桶);最后,按照众数投票确定每个桶中的最终编码。仿真结果表明在0.10和0.05错误率条件下,平均解码准确率在20X测序深度时可达94%以上;在0.15错误率条件下,平均解码准确率在60X测序深度时可达90%以上。Abstract: Compared with traditional storage, the difficulty of DeoxyriboNucleic Acid (DNA) data storage is that insertion and deletion errors in sequenced reads pose a great challenge to data recovery. For forward error-correcting coded DNA storage with one-base error-correcting capability, a bucket allocation strategy is proposed to improve the decoding accuracy and efficiency. Firstly, all identifiable DNA codes of reads in each cluster are searched and the corresponding valid codes according to the one-base error-correcting capability are determined; Then, for each identifiable DNA code, appropriate coding position (i.e. bucket) according is allocated to its position in a read; Finally, the consensus code for each bucket is determined using majority voting strategy. Simulation results show that the proposed method can correct more than 94% errors at the coverage of 20X when error rate is 5% or 10%, and correct more than 90% errors at the coverage of 60X when error rate is 15%.
-
表 1 编码表
DNA编码 字母 标点符号 数字 DNA编码 字母 标点符号 数字 TAACCG a @ 4 ACACAC l 6 TAAGGC p ¥ ACTCTG i " 5 ATCACG e $ 2 TCAGAG j % ATGAGC y , 8 ACAGGT x }{ ATGGAG g * ACTGCA f ~ 9 TACCAC k / AGACCT s + 0 ATCCGT b () TCTCGT h - ATGCCA v : TCGAAC c ? 1 TAGCGA r ' 3 ACGACT o ! TAGGCT t [] CATTCG z & ACATCG w ; CTACAG q = ACTTGC n . CAACGT d __ TCTACG m Enter CAGACA u # 7 TGCATA 大写键 GTATGA 标点符号键 CTTGTC 数字键 CGGTAT 空格键 -
[1] REINSEL D, GANTZ J, and RYDNING J. The digital of the world from edge to core[EB/OL]. http://book.itep.ru/depository/dig_economy/idc-seagate-dataage-whitepaper.pdf, 2020. [2] WILLIAMS E D, AYRES R U, and HELLER M. The 1.7 kilogram microchip: Energy and material use in the production of semiconductor devices[J]. Environmental Science & Technology, 2002, 36(24): 5504–5510. doi: 10.1021/es025643o [3] GODA K and KITSUREGAWA M. The history of storage systems[J]. Proceedings of the IEEE, 2012, 100: 1433–1440. doi: 10.1109/JPROC.2012.2189787 [4] 许鹏, 方刚, 石晓龙, 等. DNA存储及其研究进展[J]. 电子与信息学报, 2020, 42(6): 1326–1331. doi: 10.11999/JEIT190863XU Peng, FANG Gang, SHI Xiaolong, et al. DNA storage and its research progress[J]. Journal of Electronics &Information Technology, 2020, 42(6): 1326–1331. doi: 10.11999/JEIT190863 [5] 刘文斌, 朱翔鸥, 王向红, 等. 一种优化DNA计算模板性能的新方法[J]. 电子与信息学报, 2008, 30(5): 1131–1135.LIU Wenbin, ZHU Xiangou, WANG Xianghong, et al. A new method to optimize the template set in DNA computing[J]. Journal of Electronics &Information Technology, 2008, 30(5): 1131–1135. [6] CEZE L, NIVALA J, and STRAUSS K. Molecular digital data storage using DNA[J]. Nature Reviews Genetics, 2019, 20(8): 456–466. doi: 10.1038/s41576-019-0125-3 [7] GAO Yanmin, CHEN Xin, QIAO Hongyan, et al. Low-bias manipulation of DNA oligo pool for robust data storage[J]. ACS Synthetic Biology, 2020, 9(12): 3344–3352. doi: 10.1021/acssynbio.0c00419 [8] DONG Yiming, SUN Fajia, PING Zhi, et al. DNA storage: Research landscape and future prospects[J]. National Science Review, 2020, 7(6): 1092–1107. doi: 10.1093/nsr/nwaa007 [9] HECKEL R, MIKUTIS G, and GRASS R N. A characterization of the DNA data storage channel[J]. Scientific Reports, 2019, 9(1): 9663. doi: 10.1038/s41598-019-45832-6 [10] STANCU M C, VAN ROOSMALEN M J, RENKENS I, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing[J]. Nature Communications, 2017, 8(1): 1326. doi: 10.1038/s41467-017-01343-4 [11] TAKAHASHI C N, NGUYEN B H, STRAUSS K, et al. Demonstration of end-to-end automation of DNA data storage[J]. Scientific Reports, 2019, 9(1): 4998. doi: 10.1038/s41598-019-41228-8 [12] KUMAR U K and UMASHANKAR B S. Improved hamming code for error detection and correction[C]. 2007 2nd International Symposium on Wireless Pervasive Computing, San Juan, USA, 2007: 1. doi: 10.1109/ISWPC.2007.342654. [13] BLAWAT M, GAEDKE K, HÜTTER I, et al. Forward error correction for DNA data storage[J]. Procedia Computer Science, 2016, 80: 1011–1022. doi: 10.1016/j.procs.2016.05.398 [14] LU Xiaozhou, JEONG J, KIM J W, et al. Error rate-based log-likelihood ratio processing for low-density parity-check codes in DNA storage[J]. Ieee Access, 2020, 8: 162892–162902. doi: 10.1109/ACCESS.2020.3021700 [15] ORGANICK L, ANG S D, CHEN Y J, et al. Random access in large-scale DNA data storage[J]. Nature Biotechnology, 2018, 36(3): 242–248. doi: 10.1038/nbt.4079 [16] ANTKOWIAK P L, LIETARD J, DARESTANI M Z, et al. Low cost DNA data storage using photolithographic synthesis and advanced information reconstruction and error correction[J]. Nature Communications, 2020, 11(1): 5345. doi: 10.1038/s41467-020-19148-3 [17] MEISER L C, ANTKOWIAK P L, KOCH J, et al. Reading and writing digital data in DNA[J]. Nature Protocols, 2020, 15(1): 86–101. doi: 10.1038/s41596-019-0244-5 [18] ERLICH Y and ZIELINSKI D. DNA Fountain enables a robust and efficient storage architecture[J]. Science, 2017, 355(6328): 950–954. doi: 10.1126/science.aaj2038 [19] JEONG J, PARK S J, KIM J W, et al. Cooperative sequence clustering and decoding for DNA storage system with fountain codes[J]. Bioinformatics, 2021, 37(19): 3136–3143. doi: 10.1093/bioinformatics/btab246 [20] PRESS W H, HAWKINS J A, JONES JR S K, et al. HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints[J]. Proceedings of the National Academy of Sciences of the United States of America, 2020, 117(31): 18489–18496. doi: 10.1073/pnas.2004821117 [21] XUE Tianbo and LAU F C M. Notice of violation of IEEE publication principles: Construction of GC-balanced DNA with deletion/insertion/mutation error correction for DNA storage system[J]. IEEE Access, 2020, 8: 140972–140980. doi: 10.1109/ACCESS.2020.3012688 [22] SONG Lifu, GENG Feng, GONG Ziyi, et al. . Robust data storage in DNA by de Bruijn graph-based decoding[J]. bioRxiv, 2022, 13(1): 5361. doi: 10.1101/2020.12.20.423642. [23] ZAN Xiangzhen, YAO Xiangyu, XU Peng, et al. A hierarchical error correction strategy for text DNA storage[J]. Interdisciplinary Sciences: Computational Life Sciences, 2022, 14(1): 141–150. doi: 10.1007/s12539-021-00476-x. [24] BORNHOLT J, LOPEZ R, CARMEAN D M, et al. A DNA-based archival storage system[J]. ACM SIGPLAN Notices, 2016, 51(4): 637–649. doi: 10.1145/2954679.2872397 [25] ZHONG Yunpeng, QI Shanshan, SHENG Fuxu, et al. A new digital information storing and reading system based on synthetic DNA[J]. Science China Life Sciences, 2018, 61(6): 733–735. doi: 10.1007/s11427-017-9131-7 [26] LEE U J, HWANG S, KIM K E, et al. DNA data storage in Perl[J]. Biotechnology and Bioprocess Engineering, 2020, 25(4): 607–615. doi: 10.1007/s12257-020-0022-9 -






 下载:
下载:
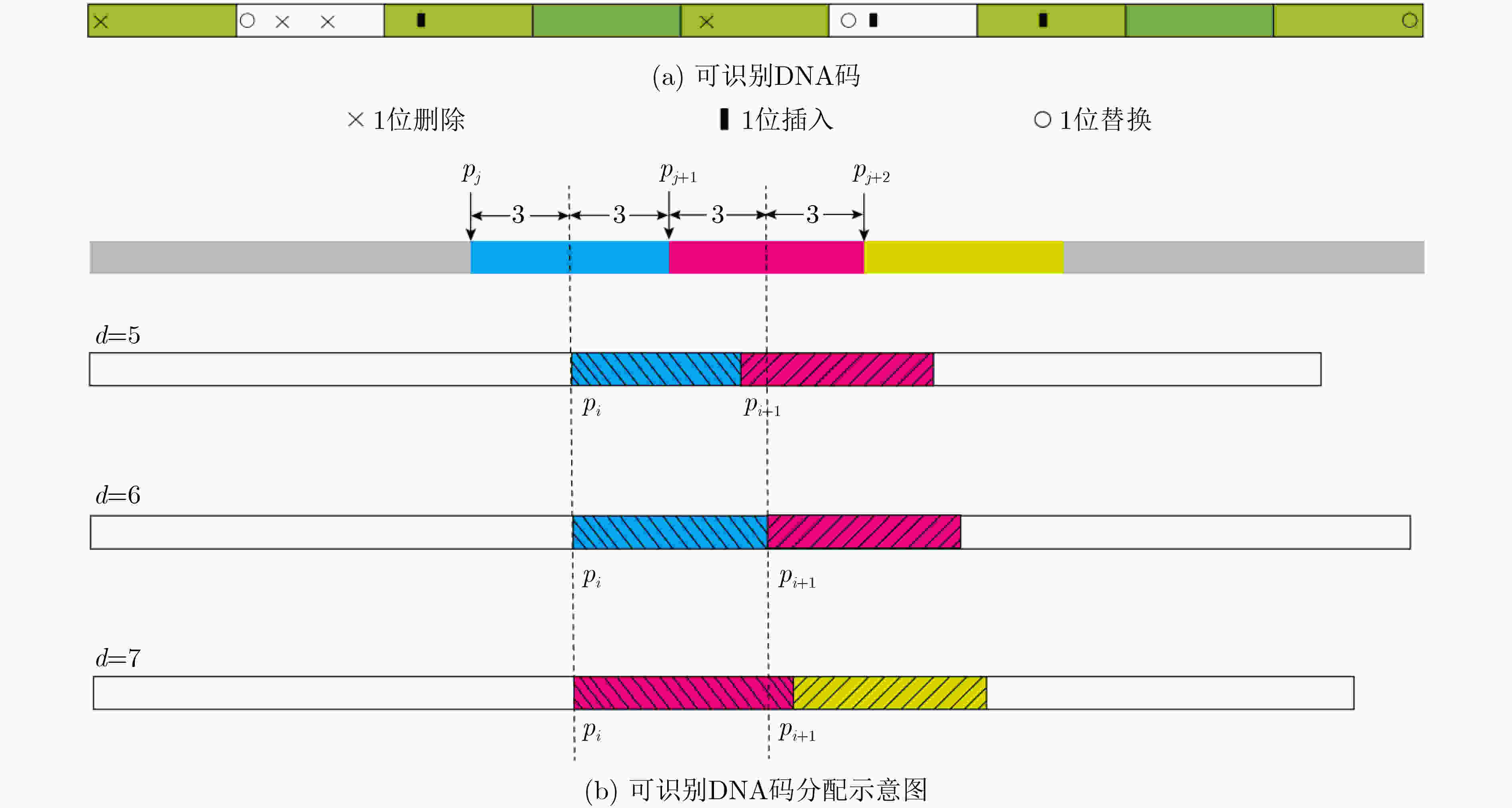
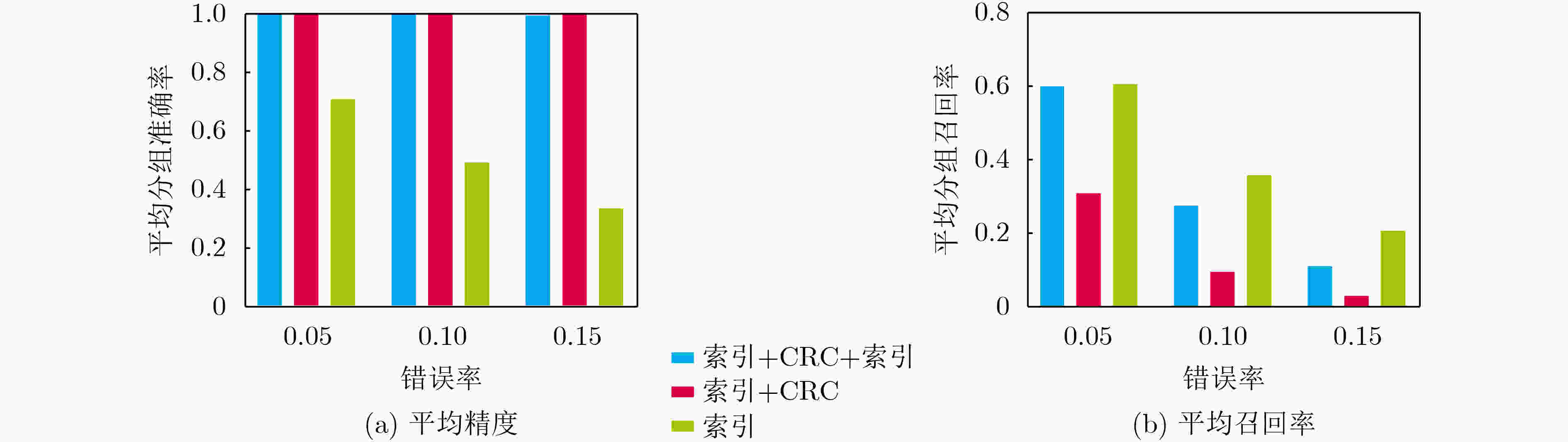
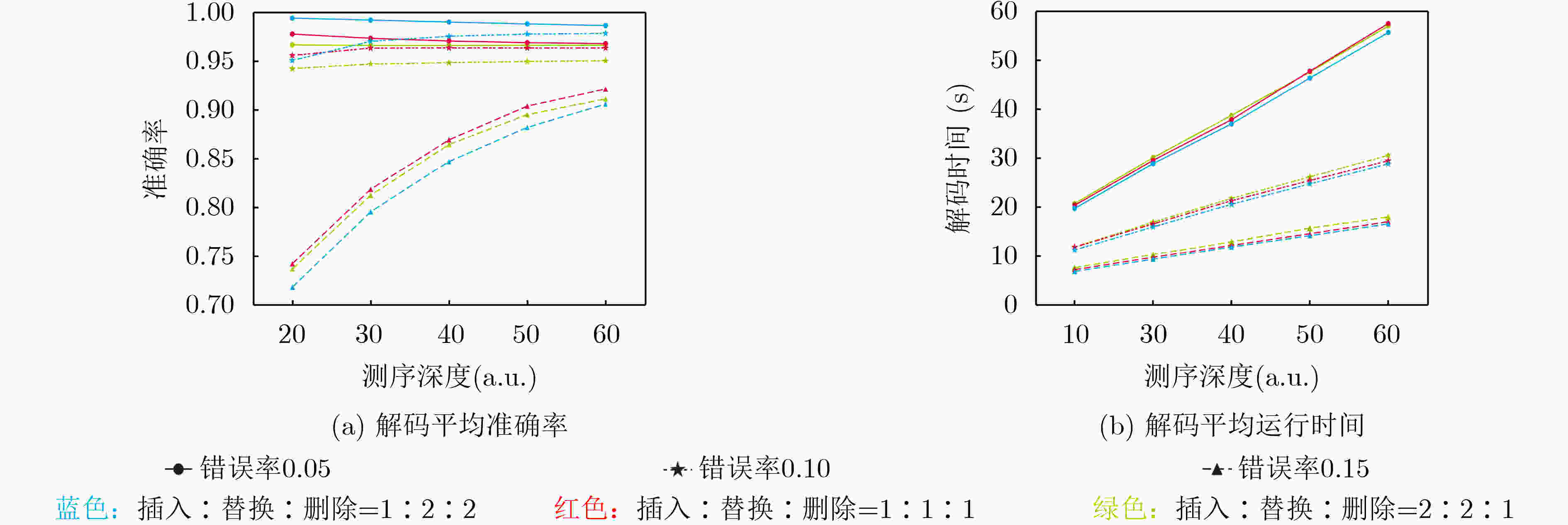


 下载:
下载:
