Automatic Liver Tumor Segmentation Based on Cascaded Dense-UNet and Graph Cuts
-
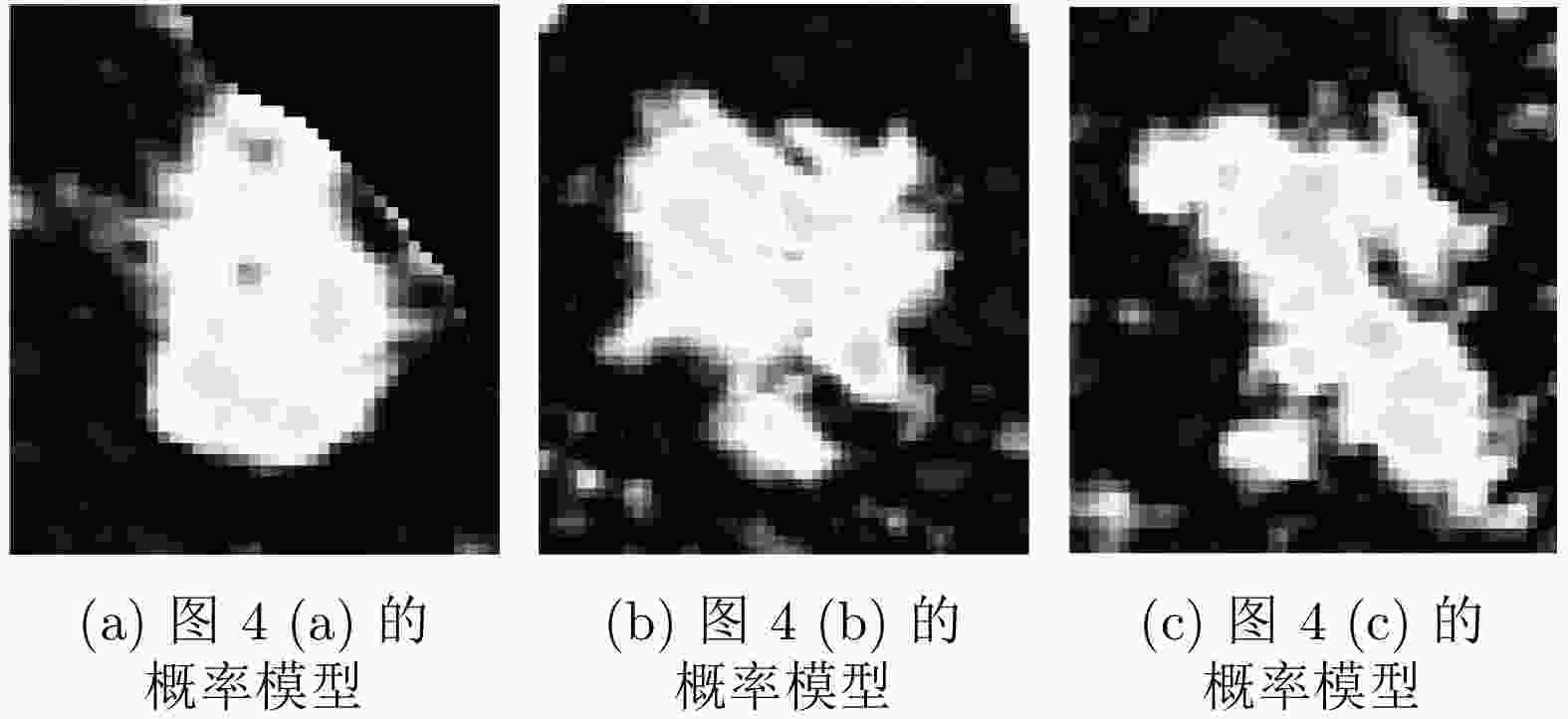
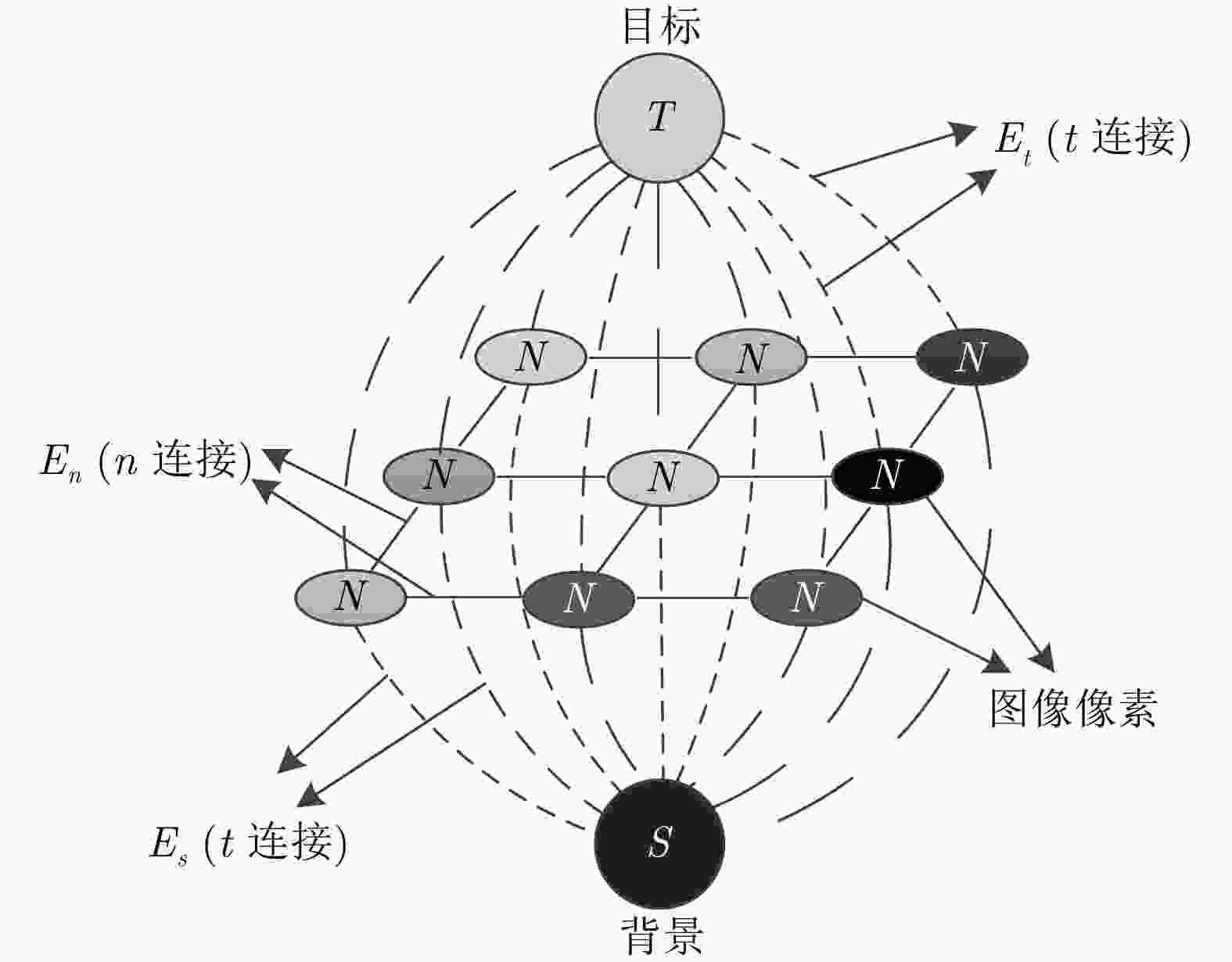
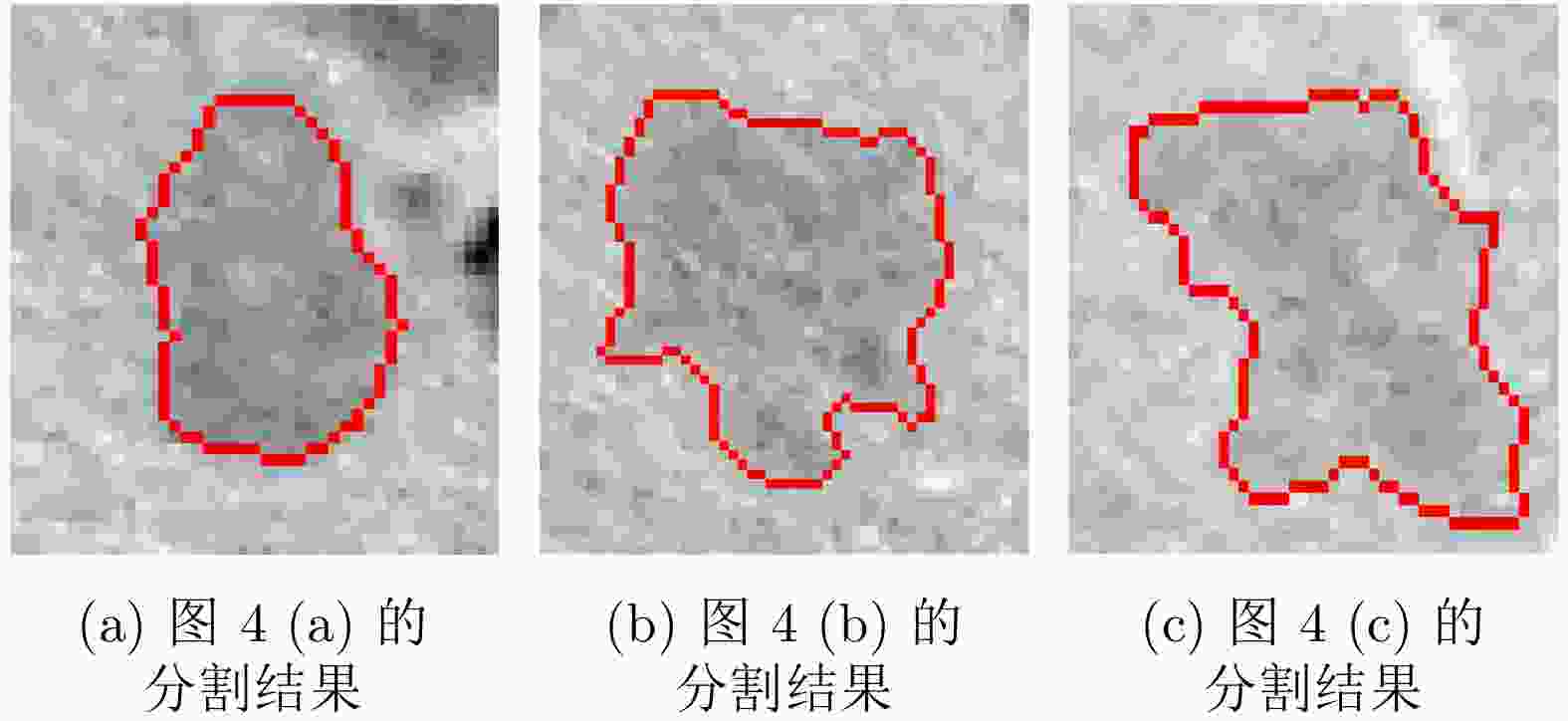
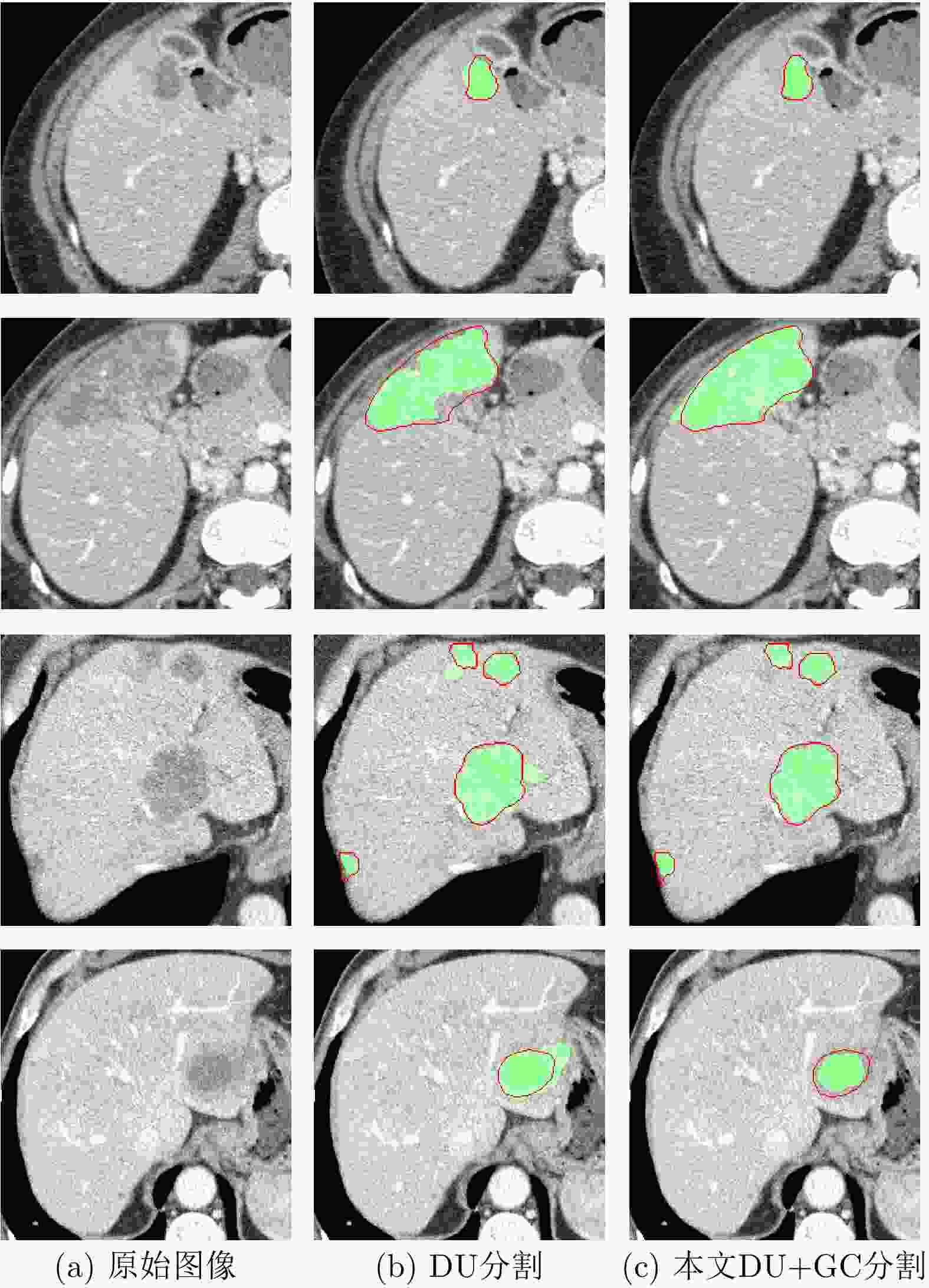
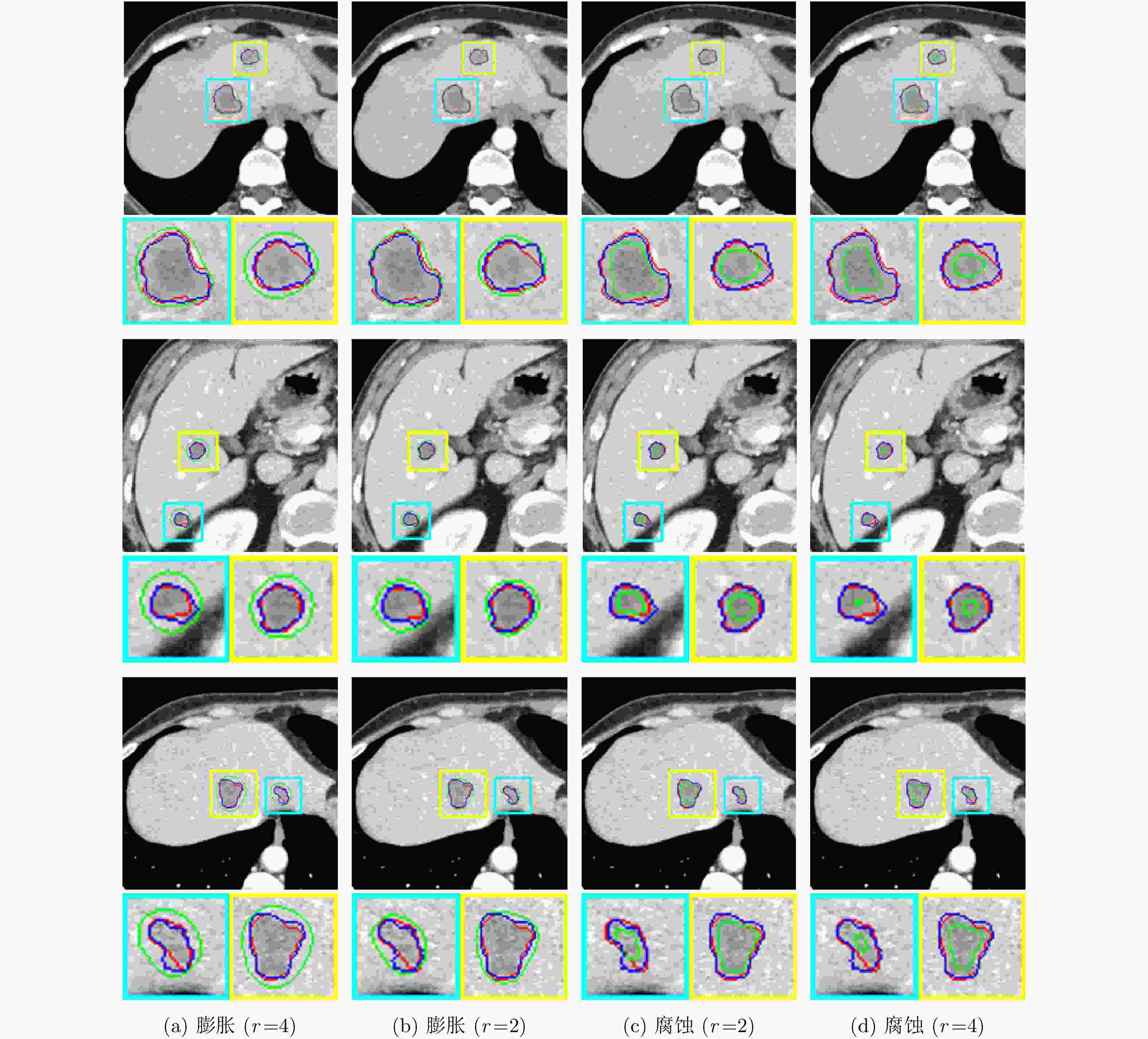
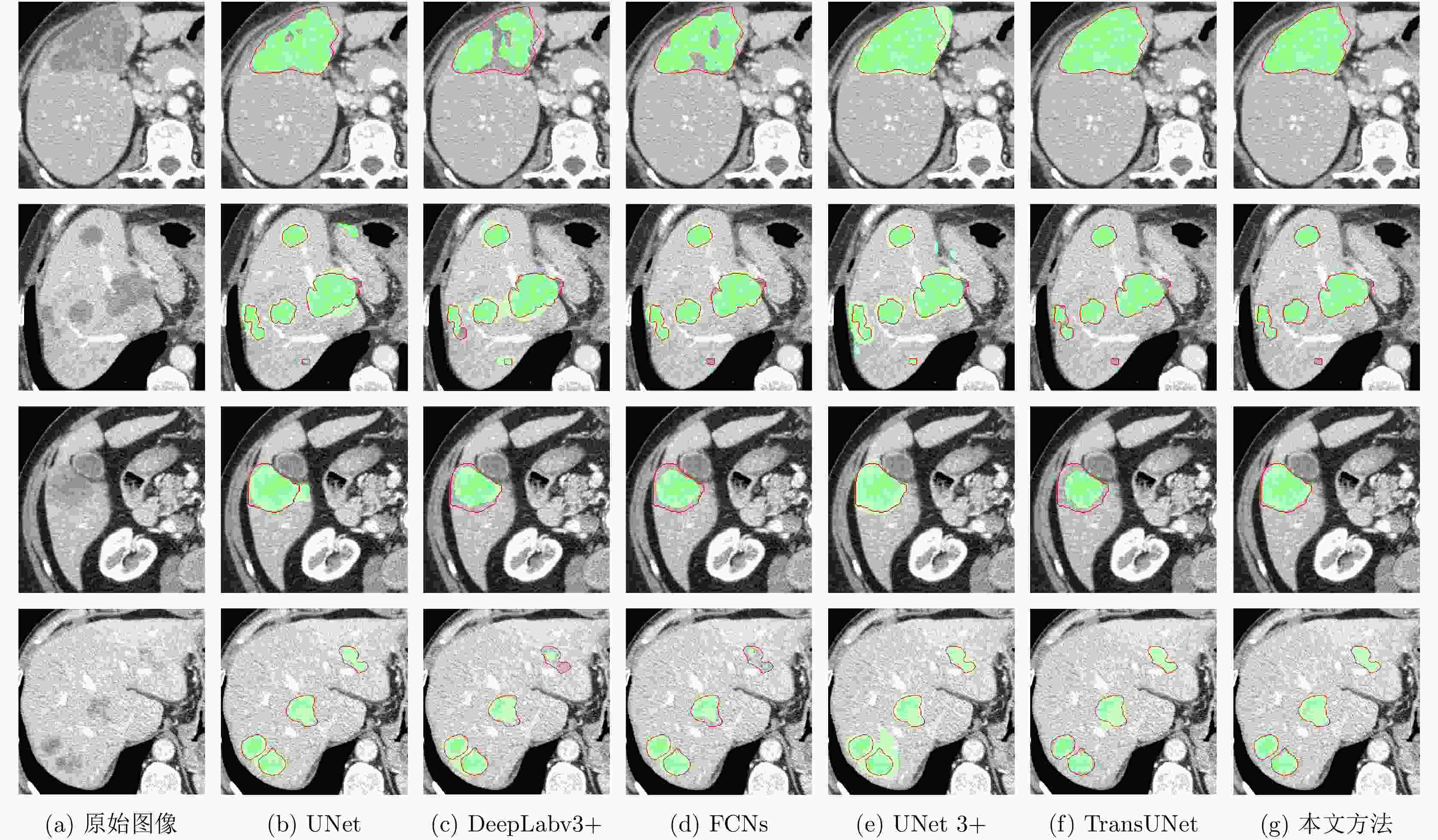
摘要: 腹部CT图像肝脏肿瘤分割是进行肝脏疾病诊断、手术规划和放射治疗的重要前提。针对肝脏肿瘤灰度异质、纹理丰富、边界模糊等因素引起的分割困难,该文提出基于级联Dense-Unet和图割的自动精确鲁棒分割方法。首先运用级联的Dense-UNet获取肝脏肿瘤初始分割结果及感兴趣区域,然后利用图像像素级和区域级特征,分别构建可有效区分肿瘤与非肿瘤的灰度模型和概率模型,并将其融入图割能量函数,进一步精确分割感兴趣区域中的肿瘤组织。最后分别采用LiTS和3Dircadb公共数据库作为训练集与测试集进行实验,并与现有多种自动分割方法进行了比较。结果表明,提出方法可有效分割CT图像中灰度、形状、大小、位置各异的肝脏肿瘤,能提取更精确的肿瘤边界,尤其对于对比度低、边界模糊的肿瘤具有明显优势。
-
关键词:
- CT图像 /
- 肿瘤分割 /
- Dense-UNet /
- 图割
Abstract: Liver tumor segmentation from abdominal CT image is an important prerequisite for liver disease diagnosis, surgical planning, and radiation therapy. However, the segmentation remains a challenging problem since the tumors in CT images generally have heterogeneous intensities, complicated textures, and ambiguous boundaries. To address this, an automatic, accurate, and robust segmentation method is proposed based on cascaded Dense-Unet and graph cuts. Firstly, the cascaded Dense-UNet is used to obtain liver tumor initial segmentation results as well as the tumor Regions Of Interest (ROIs). Then, an intensity model and a probability model are established respectively by utilizing pixel-wise and patch-wise features in order to distinguish between tumor and non-tumor, and these models are further integrated into the graph cuts energy function to segment the tumor from ROIs accurately. Finally, experiments are carried out on LiTS and 3Dircadb datasets, which are respectively used as training set and testing set, and this method is compared with many other existing automatic segmentation methods. Results demonstrate that the proposed method can segment liver tumors in CT images with different intensity, texture, shape and size more effectively and can extract the tumor boundaries more accurately than other methods, especially for the tumors with low contrasts and ambiguous boundaries.-
Key words:
- CT image /
- Tumor segmentation /
- Dense-UNet /
- Graph cuts
-
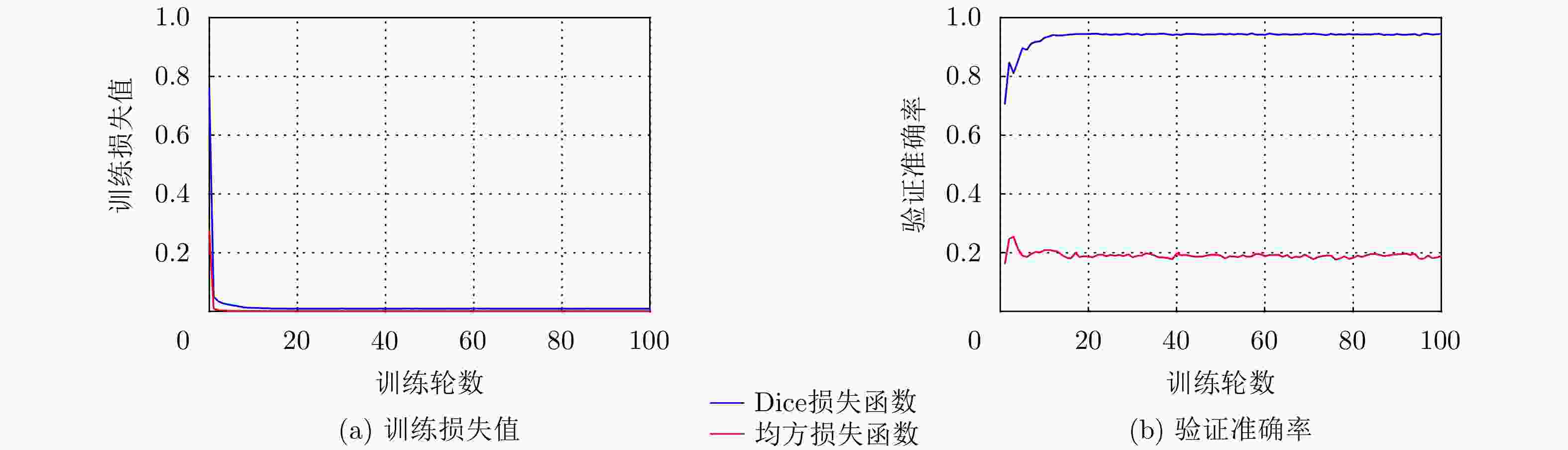
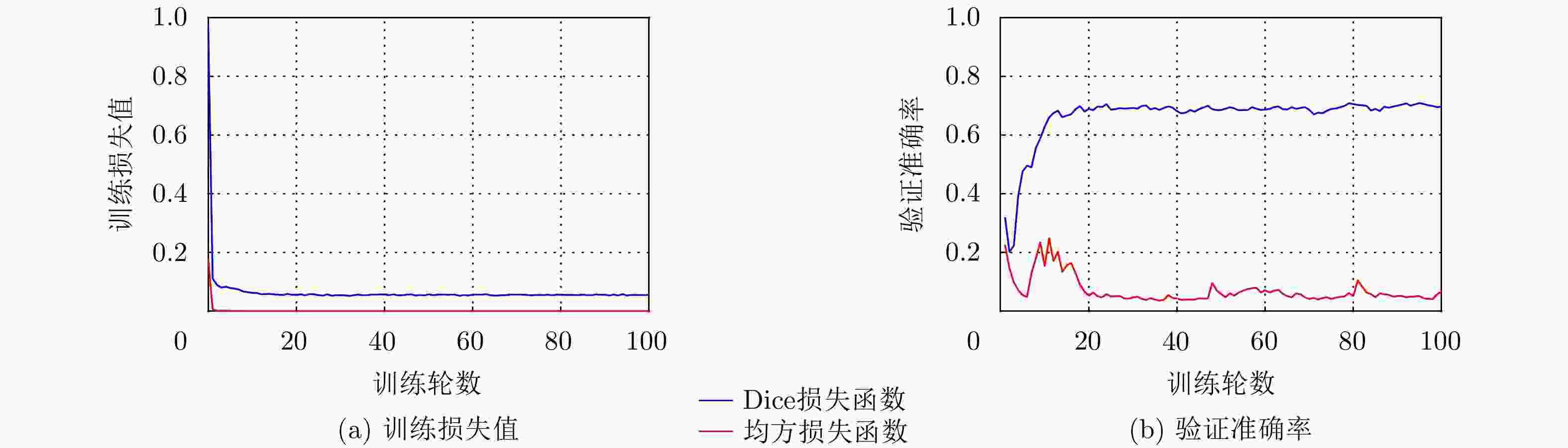
表 1 本文方法(DU+GC)与DU的分割性能比较(均值±标准差)(%)
方法 Dice Precision Recall DU 70±12 84±19 70±8 DU+GC 78±8 89±10 71±7 -
[1] CAO Wei, CHEN Hongda, YU Yiwei, et al. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020[J]. Chinese Medical Journal, 2021, 134(7): 783–791. doi: 10.1097/CM9.0000000000001474 [2] LI Yang, ZHAO Yuqian, ZHANG Fan, et al. Liver segmentation from abdominal CT volumes based on level set and sparse shape composition[J]. Computer Methods and Programs in Biomedicine, 2020, 195: 105533. doi: 10.1016/j.cmpb.2020.105533 [3] ALIRR O I. Deep learning and level set approach for liver and tumor segmentation from CT scans[J]. Journal of Applied Clinical Medical Physics, 2020, 21(10): 200–209. doi: 10.1002/acm2.13003 [4] SIRIAPISITH T, KUSAKUNNIRAN W, and HADDAWY P. Pyramid graph cut: Integrating intensity and gradient information for grayscale medical image segmentation[J]. Computers in Biology and Medicine, 2020, 126: 103997. doi: 10.1016/j.compbiomed.2020.103997 [5] KADOURY S, VORONTSOV E, and TANG An. Metastatic liver tumour segmentation from discriminant Grassmannian manifolds[J]. Physics in Medicine & Biology, 2015, 60(16): 6459–6478. doi: 10.1088/0031-9155/60/16/6459 [6] FORUZAN A H and CHEN Yenwei. Improved segmentation of low-contrast lesions using sigmoid edge model[J]. International Journal of Computer Assisted Radiology and Surgery, 2016, 11(7): 1267–1283. doi: 10.1007/s11548-015-1323-x [7] LIU Liangliang, WU Fangxiang, WANG Yuping, et al. Multi-receptive-field CNN for semantic segmentation of medical images[J]. IEEE Journal of Biomedical and Health Informatics, 2020, 24(11): 3215–3225. doi: 10.1109/jbhi.2020.3016306 [8] ISENSEE F, JAEGER P F, KOHL S A A, et al. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation[J]. Nature Methods, 2021, 18(2): 203–211. doi: 10.1038/s41592-020-01008-z [9] LI Xiaomeng, CHEN Hao, QI Xiaojuan, et al. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes[J]. IEEE Transactions on Medical Imaging, 2018, 37(12): 2663–2674. doi: 10.1109/tmi.2018.2845918 [10] JIANG Huiyan, SHI Tianyu, BAI Zhiqi, et al. AHCNet: An application of attention mechanism and hybrid connection for liver tumor segmentation in CT volumes[J]. IEEE Access, 2019, 7: 24898–24909. doi: 10.1109/access.2019.2899608 [11] LI Guoqing, ZHANG Meng, LI Jiaojie, et al. Efficient densely connected convolutional neural networks[J]. Pattern Recognition, 2021, 109: 107610. doi: 10.1016/j.patcog.2020.107610 [12] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. The 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [13] LIAO Miao, ZHAO Yuqian, LIU Xiyao, et al. Automatic liver segmentation from abdominal CT volumes using graph cuts and border marching[J]. Computer Methods and Programs in Biomedicine, 2017, 143: 1–12. doi: 10.1016/j.cmpb.2017.02.015 [14] BILIC P, CHRIST P F, VORONTSOV E, et al. The Liver Tumor Segmentation benchmark (LiTS)[EB/OL]. https://arxiv.org/abs/1901.04056, 2019. [15] SOLER L, HOSTETTLER A, AGNUS V, et al. 3D image reconstruction for comparison of algorithm database: A patient-specific anatomical and medical image database[EB/OL]. https://www.ircad.fr/research/3dircadb/, 2019. [16] SUN Changjian, GUO Shuxu, ZHANG Huimao, et al. Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs[J]. Artificial Intelligence in Medicine, 2017, 83: 58–66. doi: 10.1016/j.artmed.2017.03.008 [17] CHEN L C, ZHU Yukun, PAPANDREOU G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation[C]. The 15th European Conference on Computer Vision, Munich, Germany, 2018: 801–818. doi: 10.1007/978-3-030-01234-2_49. [18] HUANG Huimin, LIN Lanfen, TONG Ruofeng, et al. UNet 3+: A full-scale connected UNet for medical image segmentation[C]. 2020 IEEE International Conference on Acoustics, Speech and Signal Processing, Barcelona, Spain, 2020: 1055–1059. doi: 10.1109/ICASSP40776.2020.9053405. [19] CHEN Jieneng, LU Yongyi, YU Qihang, et al. TransUNet: Transformers make strong encoders for medical image segmentation[EB/OL].https://arxiv.org/abs/2102.04306, 2021. -





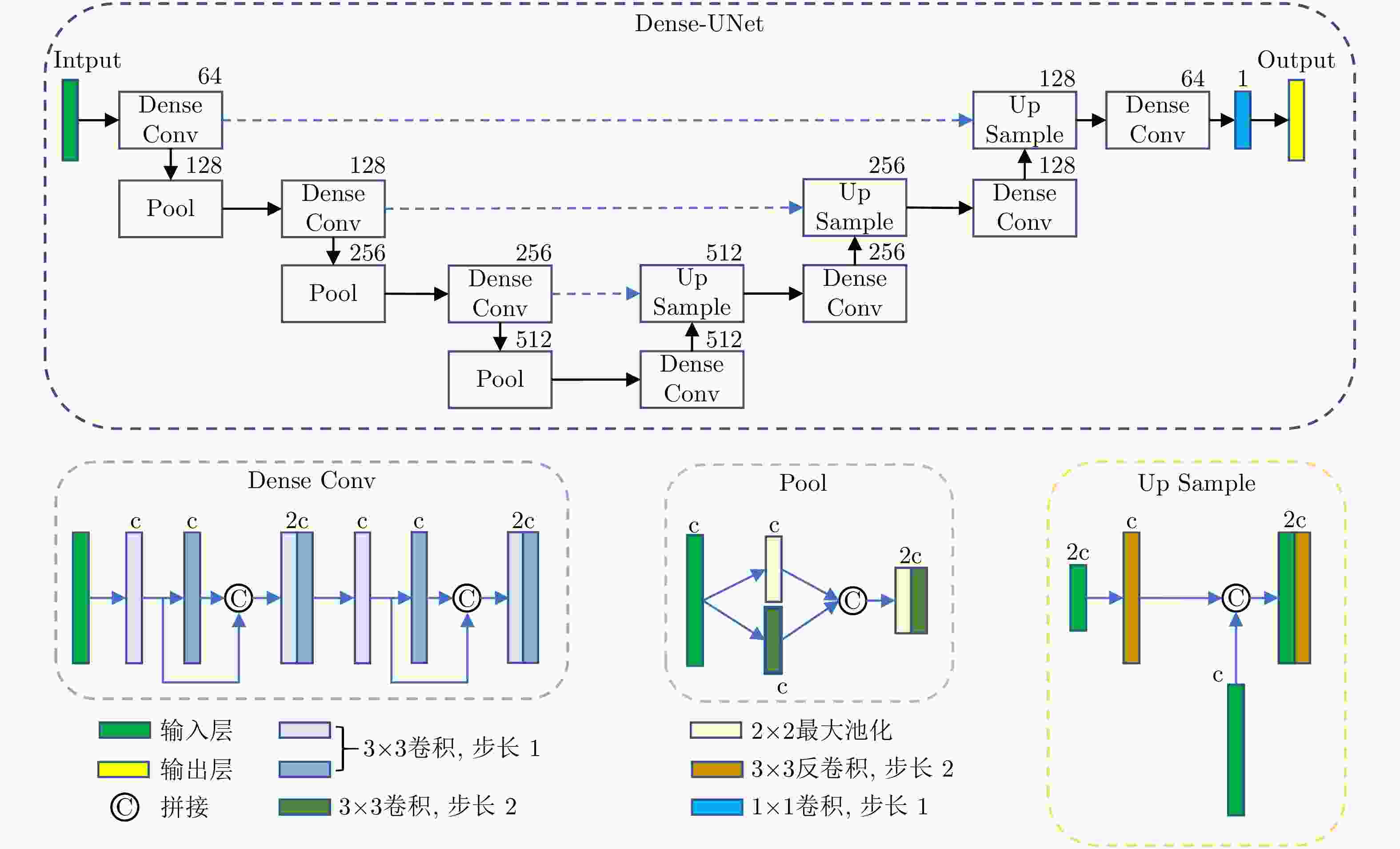
 下载:
下载:

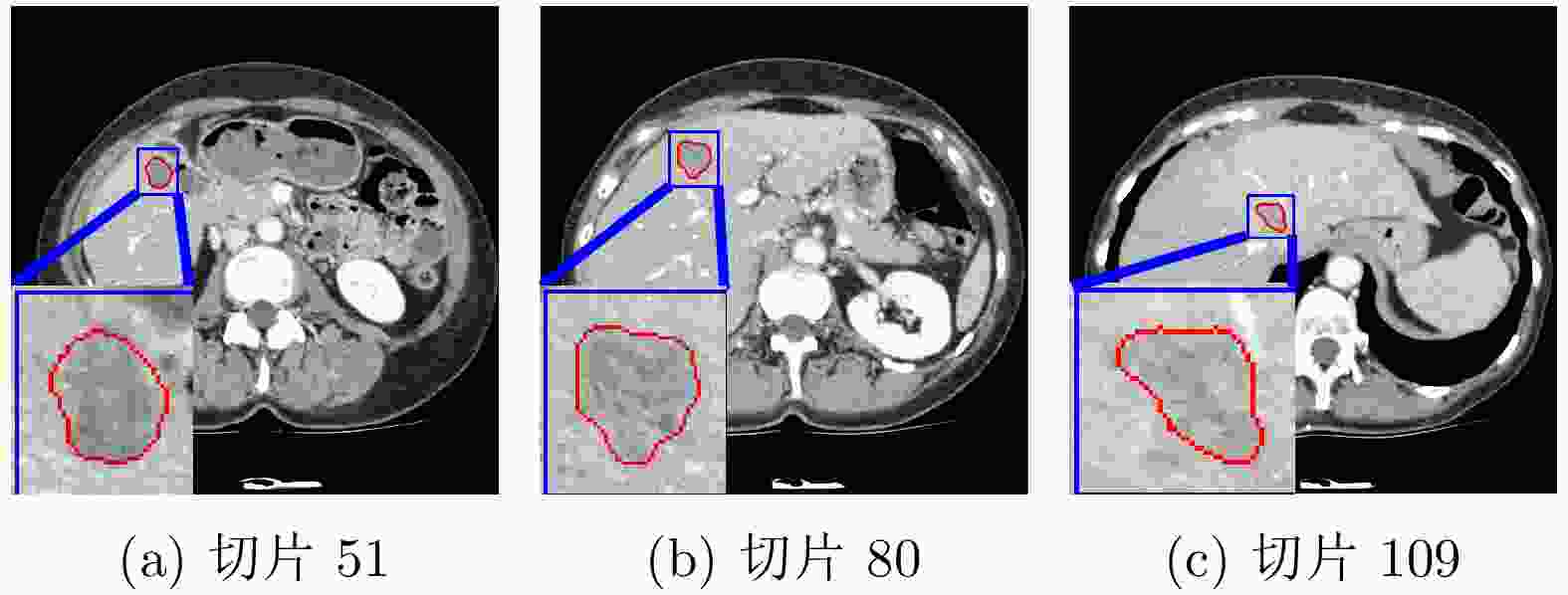
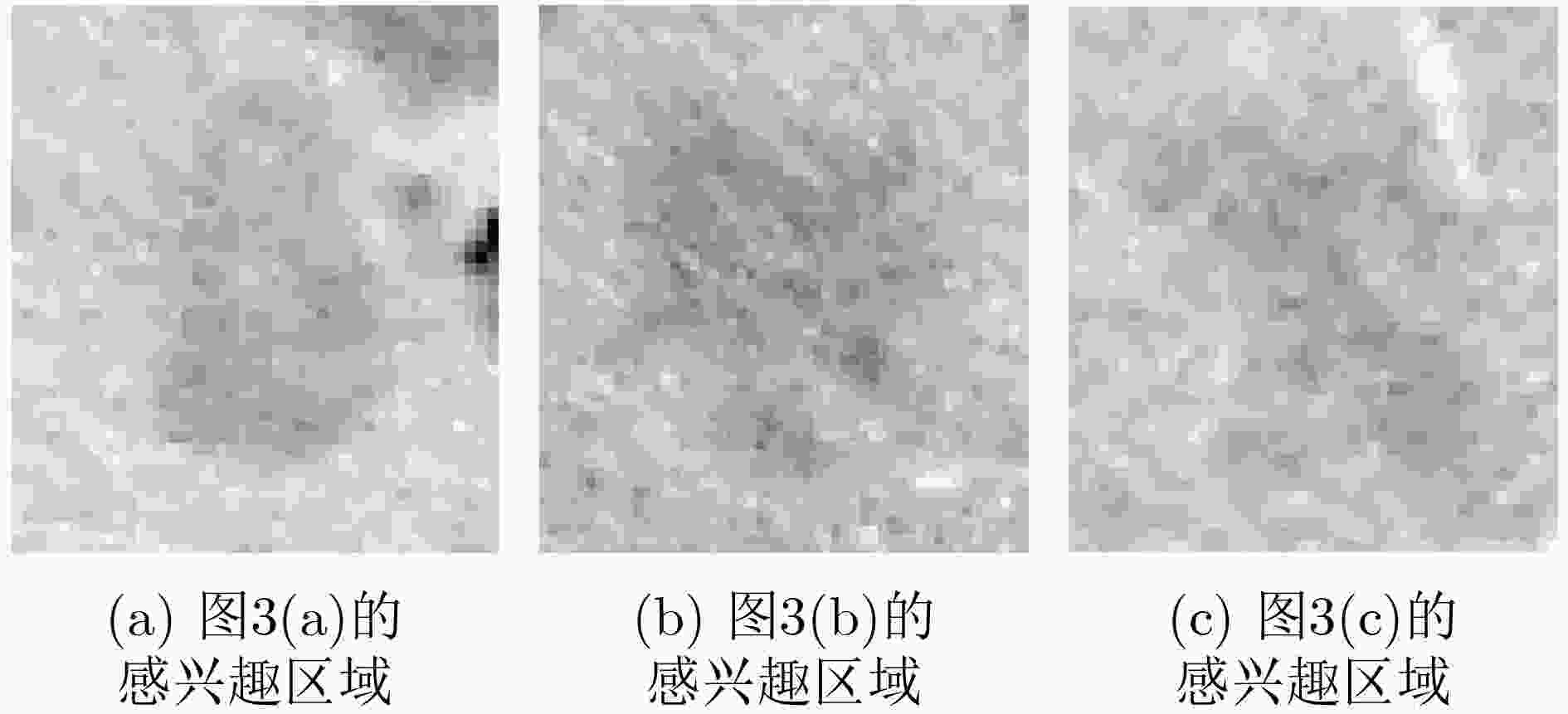
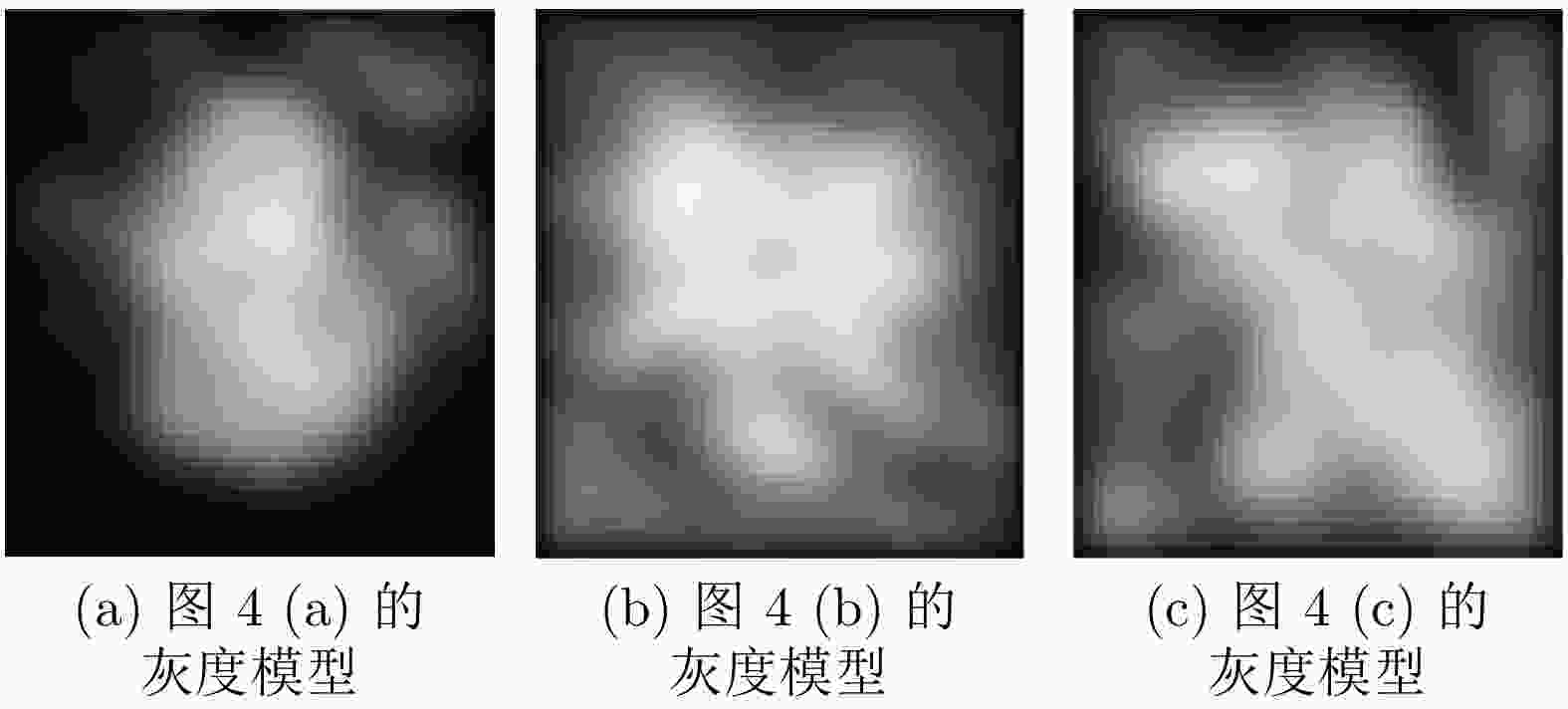


 下载:
下载:
