Computed-Tomography Image Segmentation of Cerebral Hemorrhage Based on Improved U-shaped Neural Network
-
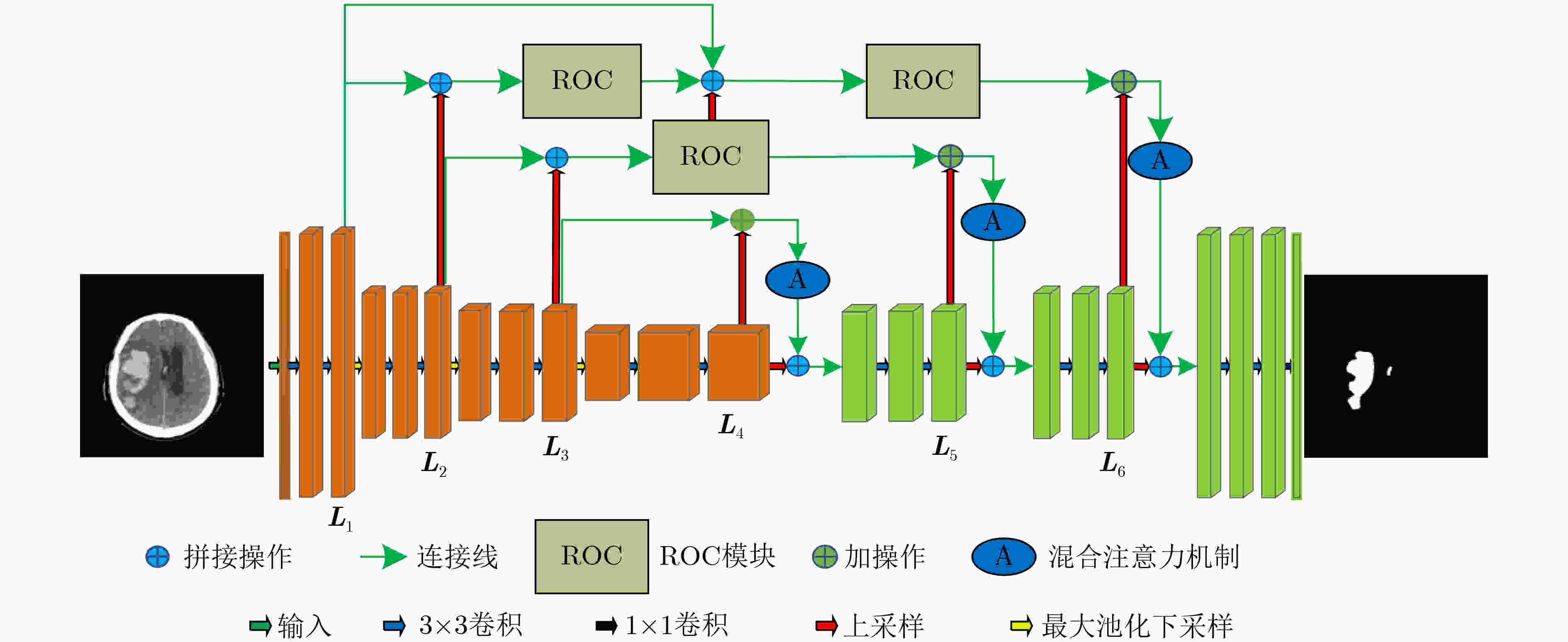
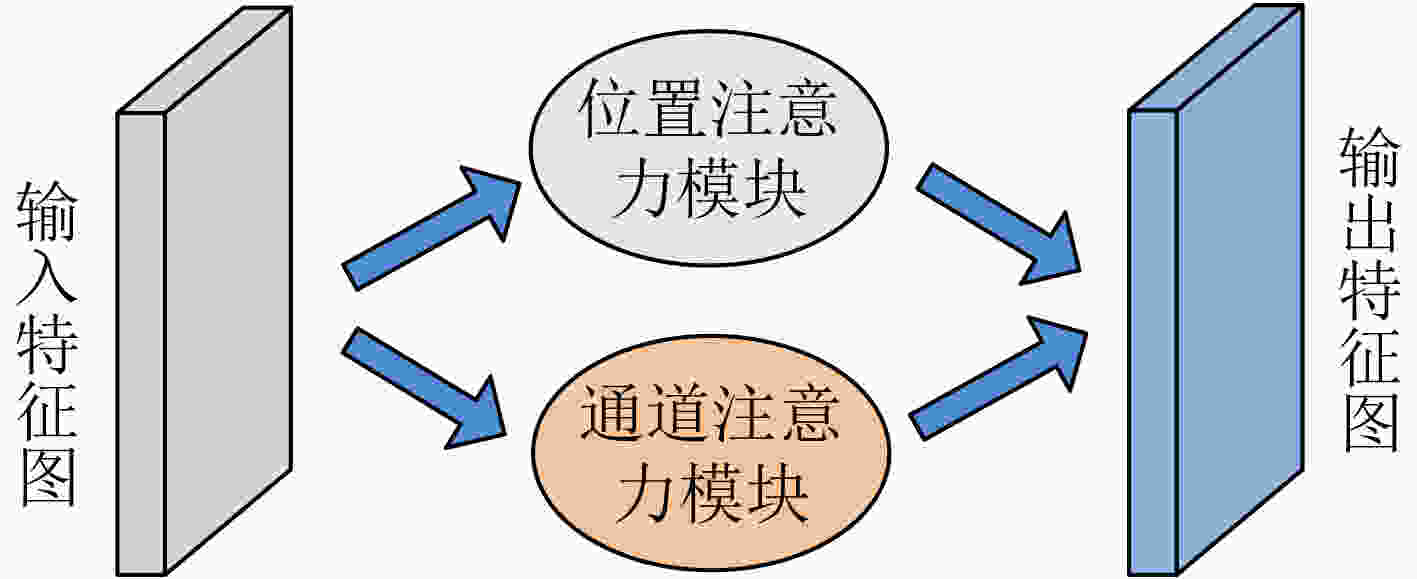
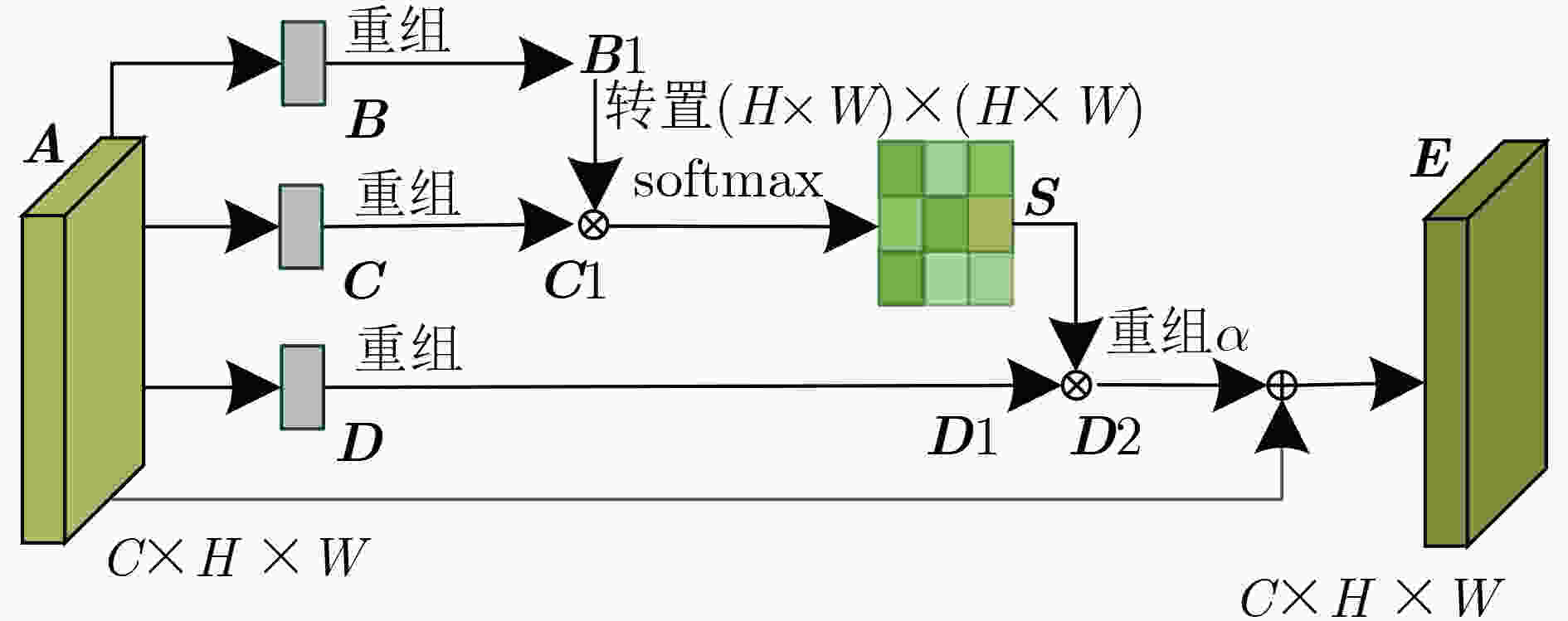
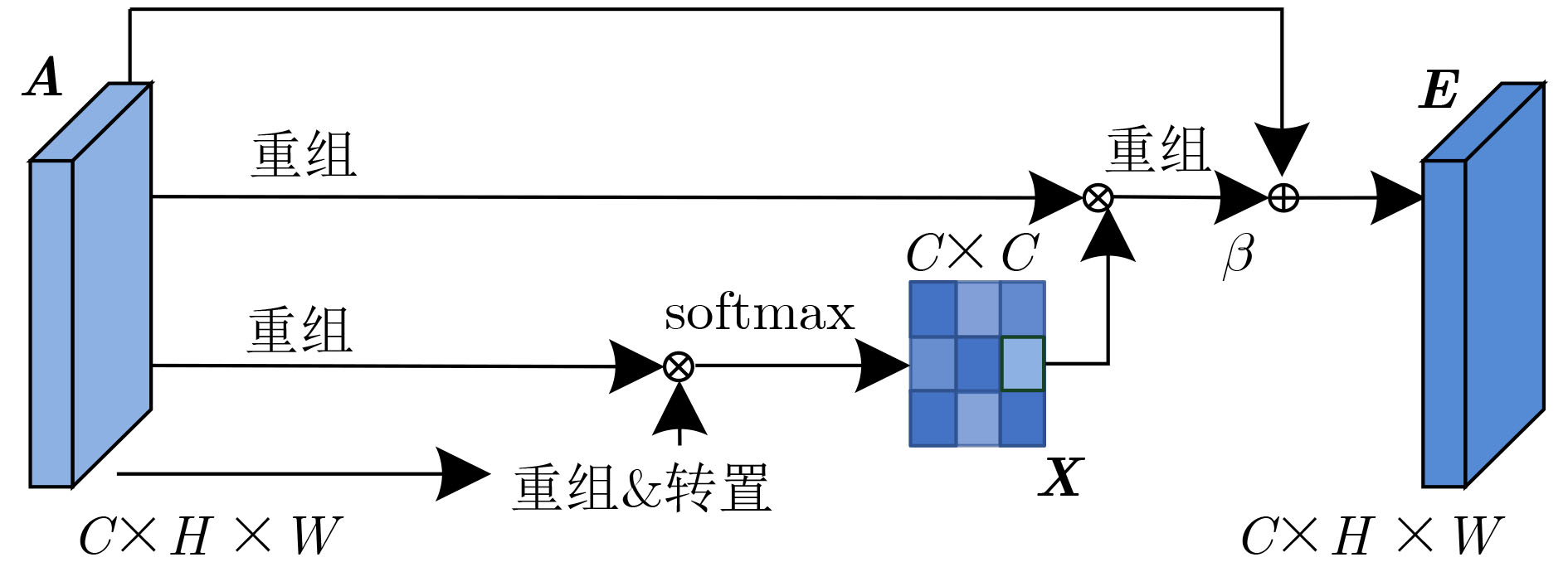
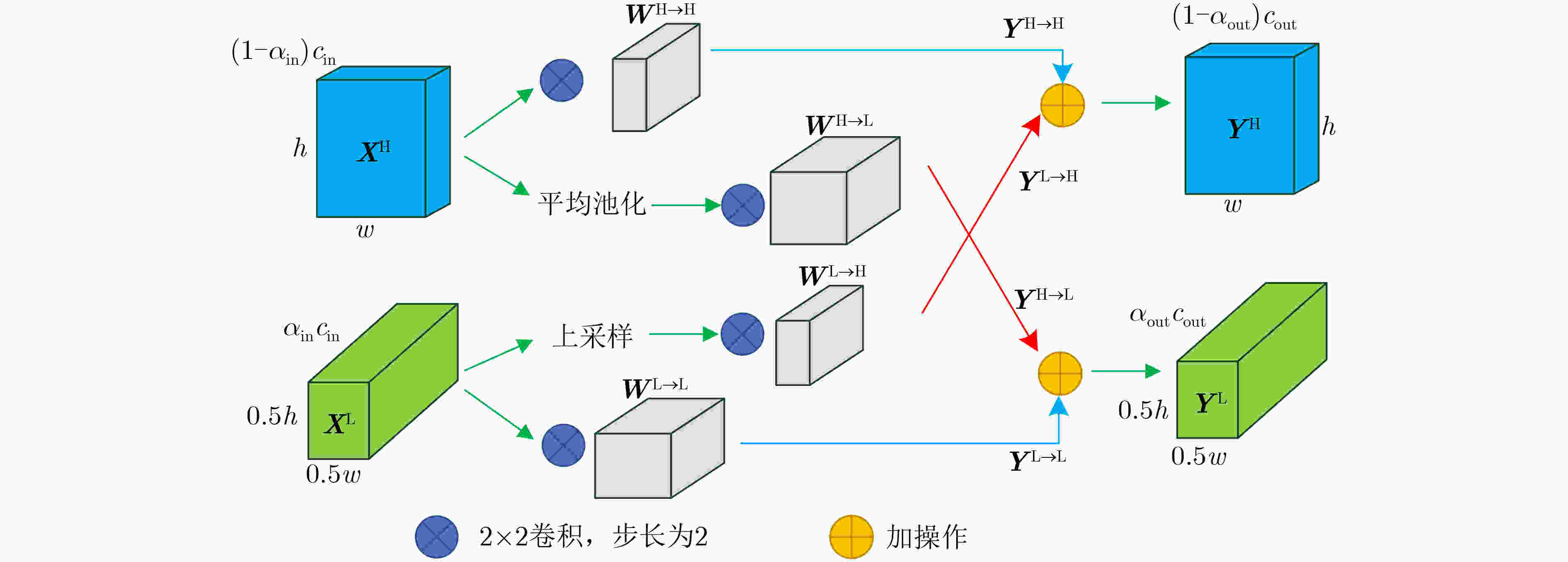
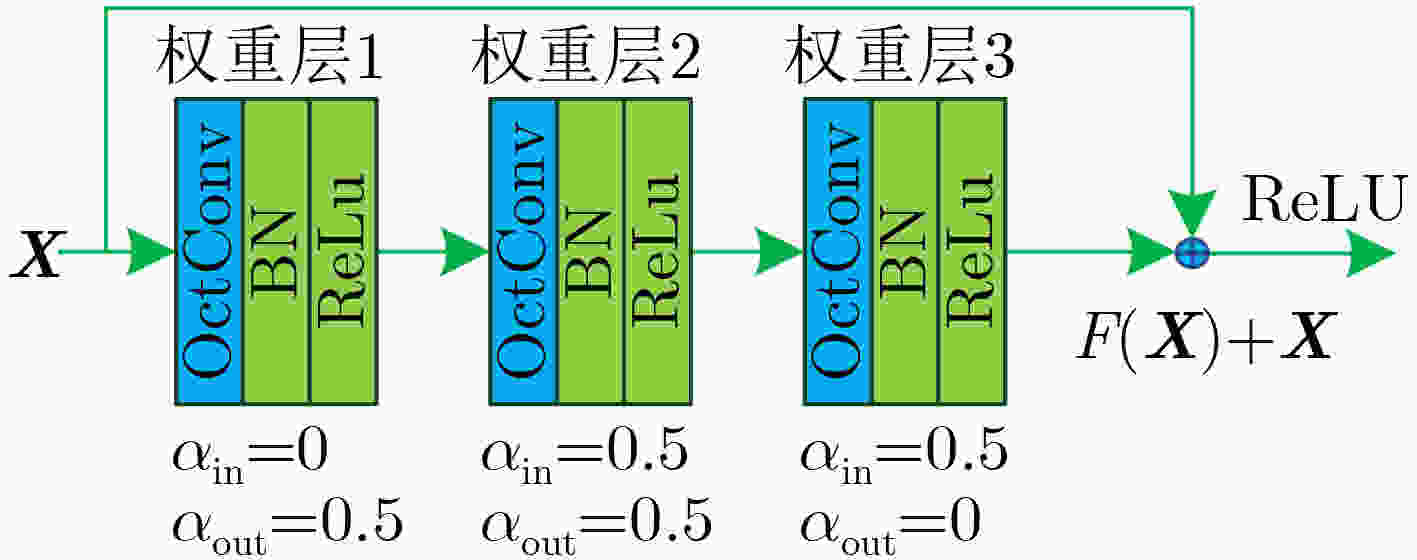
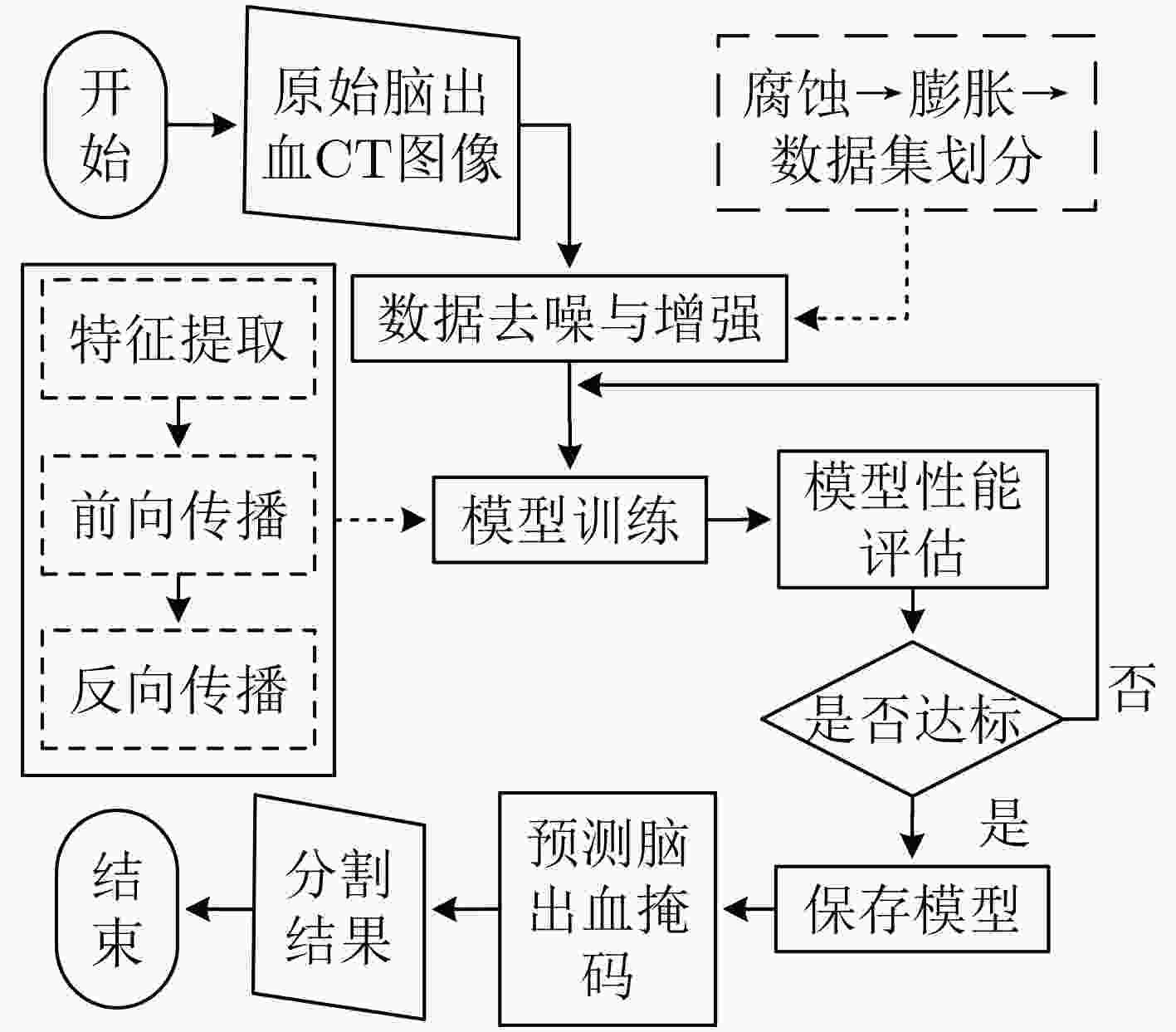
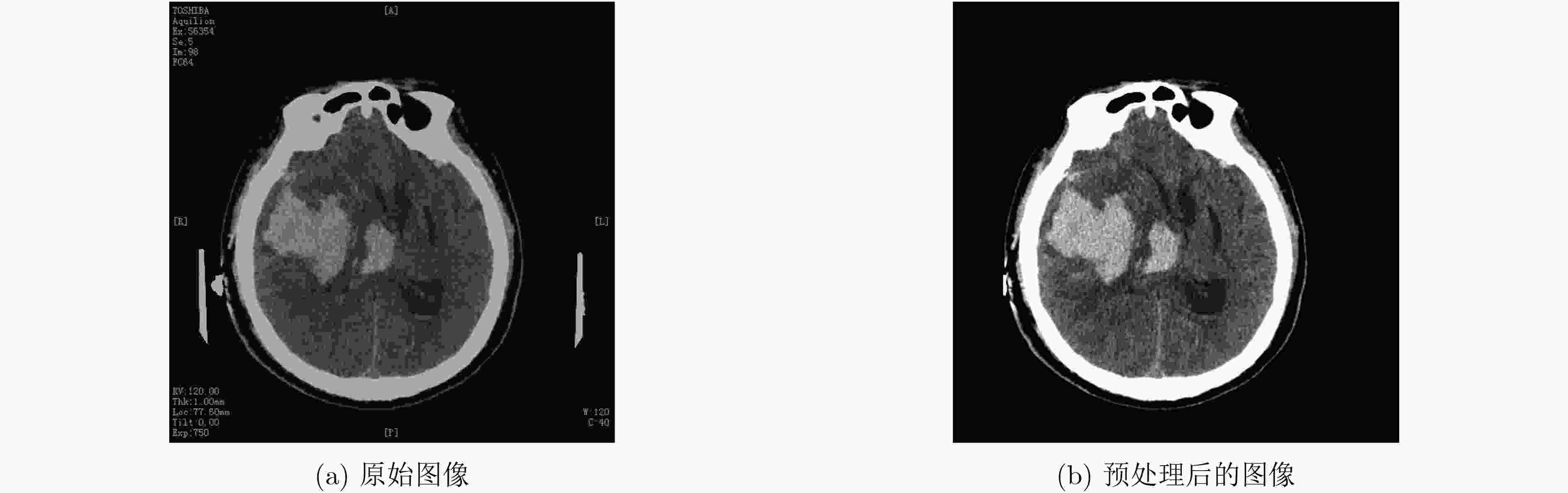
摘要: 针对脑出血CT图像病灶部位的多尺度性导致分割精度较低的问题,该文提出一种基于改进U型神经网络的图像分割模型(AU-Net+)。首先,该模型利用U-Net中的编码器对脑出血CT图像特征编码,将提出的残差八度卷积(ROC)块应用到U型神经网络的跳跃连接部分,使不同层次的特征更好地融合;其次,对融合后的特征,分别引入混合注意力机制,用以提高对目标区域的特征提取能力;最后,通过改进Dice损失函数进一步加强模型对脑出血CT图像中小目标区域的特征学习力度。为验证模型的有效性,在脑出血CT图像数据集上进行实验,同U-Net, Attention U-Net, UNet++以及CE-Net相比,mIoU指标分别提升了20.9%, 3.6%, 7.0%, 3.1%,表明AU-Net+模型具有更好的分割效果。Abstract: In view of the problem of low segmentation accuracy caused by the multi-scale of the lesion location in Computed-Tomography (CT) images of cerebral hemorrhage, an image segmentation model based on Attention improved U-shaped neural Network plus (AU-Net+) is proposed. Firstly, the model uses the encoder in U-Net to encode the features of the CT image of cerebral hemorrhage, and applies the proposed Residual Octave Convolution (ROC) block to the jump connection part of the U-shaped neural network to make the features of different levels more blend well. Secondly, for the merged features, a hybrid attention mechanism is introduced to improve the feature extraction ability of the target area. Finally, the Dice loss function is improved to enhance further the feature learning of the model for small and medium-sized target regions in CT images of cerebral hemorrhage. To verify the performance of the model, the mIoU index is improved by 20.9%, 3.6%, 7.0%, 3.1% compared with U-Net, Attention U-Net, UNet++ and CE-Net respectively, which indicates that AU-Net+ model has better segmentation effect.
-
表 1 AU-Net+网络结构
编码器-解码器 跳跃连接 conv2d_1 (UConv2D) up_sampling2d_4 (Conv2DTrans) max_pooling2d_1 (MaxPooling2D) concatenate_4 (Concatenate) conv2d_2 (UConv2D) roc_1(Roc) max_pooling2d_2 (MaxPooling2D) up_sampling2d_5 (Conv2DTrans) conv2d_3 (UConv2D) concatenate_5 (Concatenate) max_pooling2d_3 (MaxPooling2D) roc_2(Roc) conv2d_4 (UConv2D) up_sampling2d_6 (Conv2DTrans) dropout_1 (Dropout) add_1 (Add) up_sampling2d_1 (Conv2DTrans) att_1(Attention) concatenate_1 (Concatenate) up_sampling2d_7 (Conv2DTrans) conv2d_5 (UConv2D) concatenate_6 (Concatenate) up_sampling2d_2 (Conv2DTrans) roc_3(Roc) concatenate_2 (Concatenate) up_sampling2d_8 (Conv2DTrans) conv2d_6 (UConv2D) add_2 (Add) up_sampling2d_3 (Conv2DTrans) att_2(Attention) concatenate_3 (Concatenate) up_sampling2d_9 (Conv2DTrans) conv2d_7 (UConv2D) add_3 (Add) conv2d_8 (EConv2D) att_3(Attention) 表 2 分类结果的混淆矩阵
预测值\实际值 正样本 负样本 正样本 ${\rm{TP}}$ ${\rm{FP}}$ 负样本 ${\rm{FN}}$ ${\rm{TN}}$ 表 3 评价指标的统计结果
评价指标 mIoU VOE Recall DICE Specificity 均值 0.862 0.021 0.912 0.924 0.987 方差 0.009 0.001 0.004 0.002 0.002 中值 0.901 0.023 0.935 0.953 0.998 表 4 实验结果对比
方法(参数量) 迭代次数 mIoU VOE Recall DICE Specificity U-Net (31377858) 4600 0.653 0.043 0.731 0.706 0.974 Attention U-Net(31901542) 4600 0.826 0.021 0.861 0.905 0.977 U-Net++(36165192) 4800 0.792 0.025 0.833 0.883 0.976 CE-Net (29003094) 4500 0.831 0.022 0.873 0.911 0.981 AU-Net+(37646416) 5000 0.862 0.021 0.912 0.924 0.987 表 5 混合注意力机制和ROC结构分析指标对比
模型 mIoU VOE Recall DICE Specificity Network_1 0.661 0.042 0.735 0.714 0.976 Network_2 0.835 0.025 0.841 0.893 0.974 Network_3 0.781 0.041 0.744 0.723 0.985 Network_4 0.842 0.023 0.862 0.905 0.986 AU-Net+ 0.862 0.021 0.912 0.924 0.987 表 6 实验结果对比
模型 参数量 mIoU VOE Recall DICE Specificity Attention U-Net* 38654416 0.804 0.027 0.853 0.896 0.956 Attention U-Net 31901542 0.826 0.021 0.861 0.905 0.977 AU-Net+ 37646416 0.862 0.021 0.912 0.924 0.987 -
[1] 谈山峰, 方芳, 陈兵, 等. 脑疝后脑梗塞预后因素分析[J]. 海南医学, 2014, 25(3): 400–402. doi: 10.3969/j.issn.1003-6350.2014.03.0152TAN Shanfeng, FANG Fang, CHEN Bing, et al. Analysis of prognostic factors of cerebral infarction after cerebral hernia[J]. Hainan Medical Journal, 2014, 25(3): 400–402. doi: 10.3969/j.issn.1003-6350.2014.03.0152 [2] SUN Mingjie, HU R, YU Huimin, et al. Intracranial hemorrhage detection by 3D voxel segmentation on brain CT images[C]. 2015 International Conference on Wireless Communications & Signal Processing (WCSP), Nanjing, China, 2015: 1–5. doi: 10.1109/WCSP.2015.7341238. [3] WANG Nian, TONG Fei, TU Yongcheng, et al. Extraction of cerebral hemorrhage and calculation of its volume on CT image using automatic segmentation algorithm[J]. Journal of Physics: Conference Series, 2019, 1187(4): 042088. doi: 10.1088/1742-6596/1187/4/042088 [4] BHADAURIA H S, SINGH A, and DEWAL M L. An integrated method for hemorrhage segmentation from brain CT Imaging[J]. Computers & Electrical Engineering, 2013, 39(5): 1527–1536. doi: 10.1016/j.compeleceng.2013.04.010 [5] SHAHANGIAN B and POURGHASSEM H. Automatic brain hemorrhage segmentation and classification in CT scan images[C]. 2013 8th Iranian Conference on Machine Vision and Image Processing (MVIP), Zanjan, Iran, 2013: 467–471. doi: 10.1109/IranianMVIP.2013.6780031. [6] KRIZHEVSKY A, SUTSKEVER I, and HINTON G E. ImageNet classification with deep convolutional neural networks[J]. Communications of the ACM, 2017, 60(6): 84–90. doi: 10.1145/3065386 [7] WANG Shuxin, CAO Shilei, WEI Dong, et al. LT-Net: Label transfer by learning reversible voxel-wise correspondence for one-shot medical image segmentation[C]. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, USA, 2020: 9159–9168. doi: 10.1109/CVPR42600.2020.00918. [8] RONNEBERGER O, FISCHER P, and BROX T. U-Net: Convolutional networks for biomedical image segmentation[C]. The 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 2015: 234–241. doi: 10.1007/978-3-319-24574-4_28. [9] 彭佳林, 揭萍. 基于序列间先验约束和多视角信息融合的肝脏CT图像分割[J]. 电子与信息学报, 2018, 40(4): 971–978. doi: 10.11999/JEIT170933PENG Jialin and JIE Ping. Liver segmentation from CT image based on sequential constraint and multi-view information fusion[J]. Journal of Electronics &Information Technology, 2018, 40(4): 971–978. doi: 10.11999/JEIT170933 [10] MILLETARI F, NAVAB N, and AHMADI S A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation[C]. 2016 Fourth International Conference on 3D Vision (3DV), Stanford, USA, 2016: 565–571. doi: 10.1109/3DV.2016.79. [11] GUAN S, KHAN A A, SIKDAR S, et al. Fully dense UNet for 2-D sparse photoacoustic tomography artifact removal[J]. IEEE Journal of Biomedical and Health Informatics, 2020, 24(2): 568–576. doi: 10.1109/JBHI.2019.2912935 [12] XIAO Xiao, LIAN Shen, LUO Zhiming, et al. Weighted Res-UNet for high-quality retina vessel segmentation[C]. 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 2018: 327–331. doi: 10.1109/ITME.2018.00080. [13] OKTAY O, SCHLEMPER J, LE FOLGOC L, et al. Attention U-Net: Learning where to look for the pancreas[C]. The 1st Conference on Medical Imaging with Deep Learning, Amsterdam, Netherlands, 2018: 1–10. [14] IBTEHAZ N and RAHMAN M S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation[J]. Neural Networks, 2020, 121: 74–87. doi: 10.1016/j.neunet.2019.08.025 [15] ALOM M Z, YAKOPCIC C, TAHA T M, et al. Nuclei segmentation with recurrent residual convolutional neural networks based U-Net (R2U-Net)[C]. NAECON 2018-IEEE National Aerospace and Electronics Conference, Dayton, USA, 2018: 228–233. doi: 10.1109/NAECON.2018.8556686. [16] ZHOU Zongwei, RAHMAN M M, TAJBAKHSH N, et al. UNet++: Redesigning skip connections to exploit multiscale features in image segmentation[J]. IEEE Transactions on Medical Imaging, 2020, 39(6): 1856–1867. doi: 10.1109/TMI.2019.2959609 [17] GU Zaiwang, CHENG Jun, FU Huazhu, et al. CE-Net: Context encoder network for 2D medical image segmentation[J]. IEEE Transactions on Medical Imaging, 2019, 38(10): 2281–2292. doi: 10.1109/TMI.2019.2903562 [18] FU Jun, LIU Jing, TIAN Haijie, et al. Dual attention network for scene segmentation[C]. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, USA, 2019: 3141–3149. doi: 10.1109/CVPR.2019.00326. [19] CHEN Yunpeng, FAN Haoqi, XU Bing, et al. Drop an Octave: Reducing spatial redundancy in convolutional neural networks with Octave convolution[C]. 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, South Korea, 2019: 3434–3443. doi: 10.1109/ICCV.2019.00353. -





 下载:
下载:
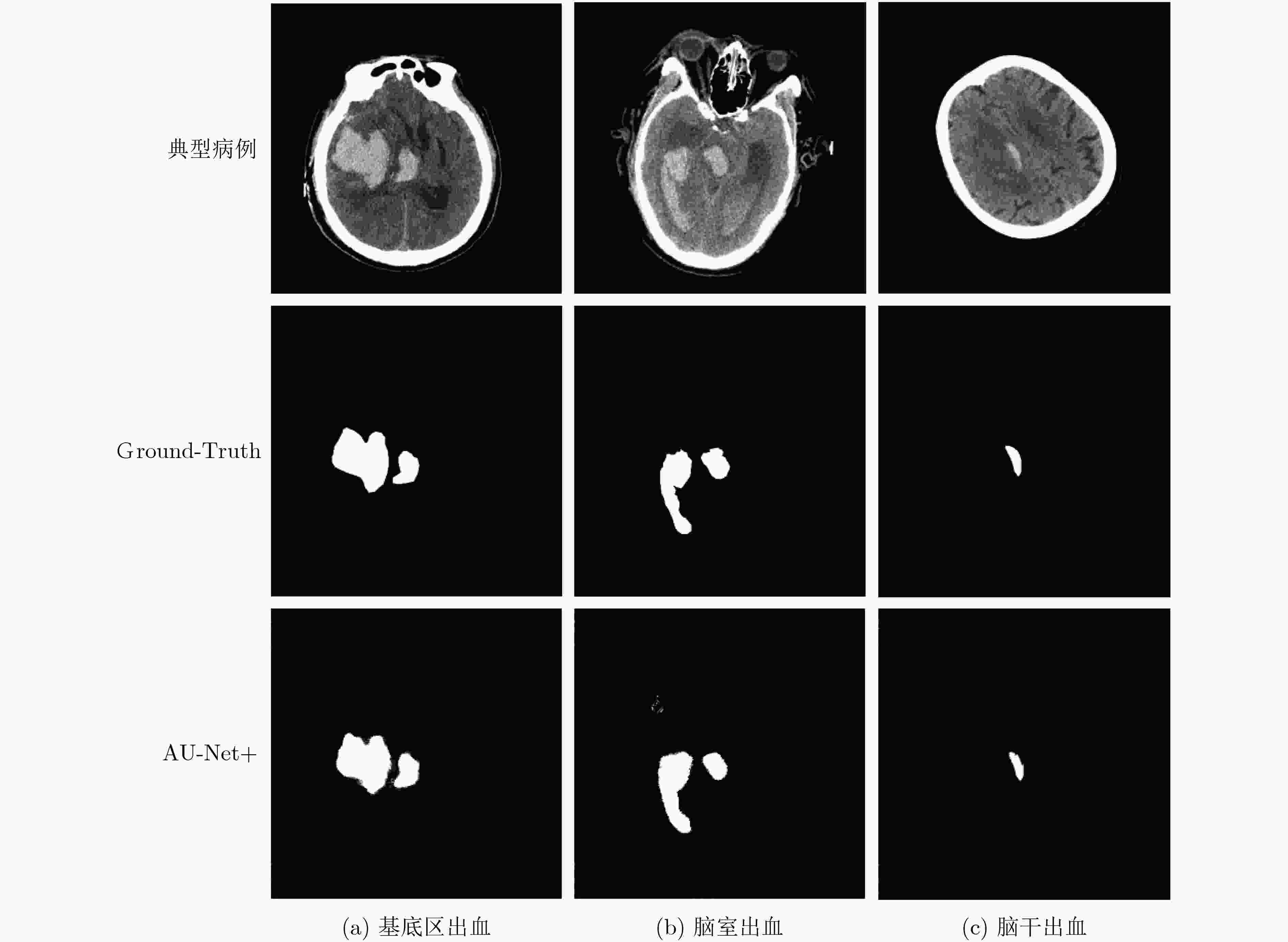
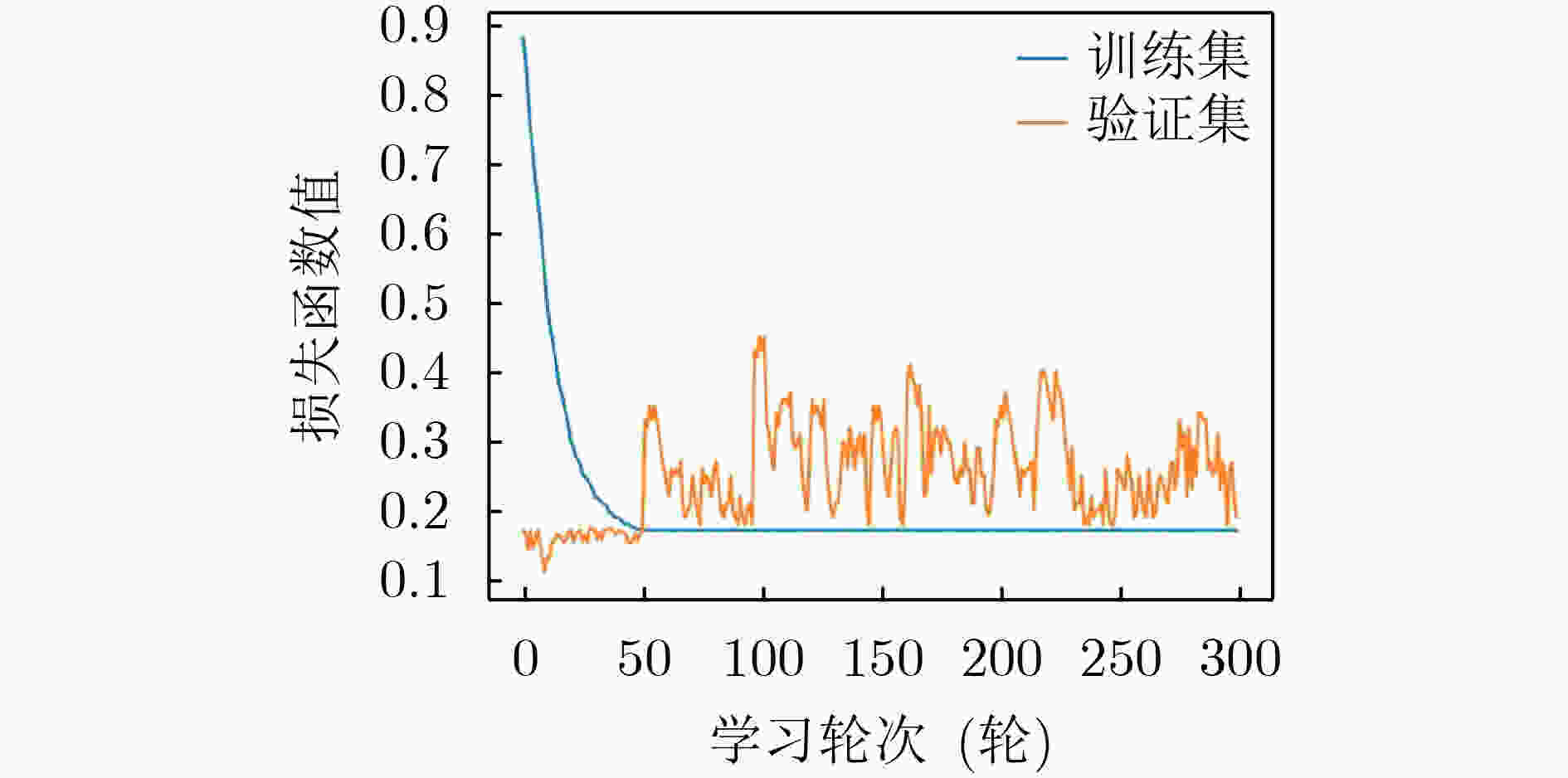

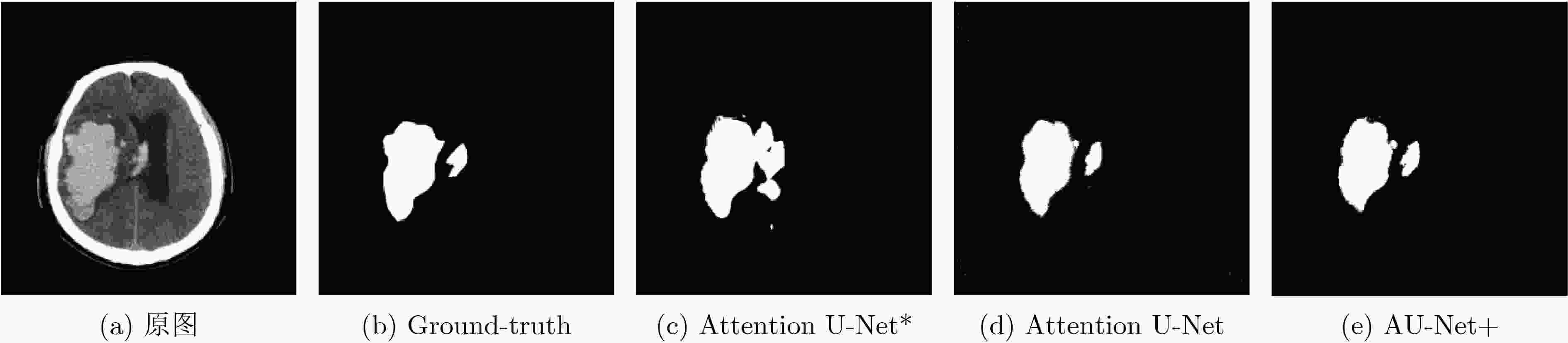
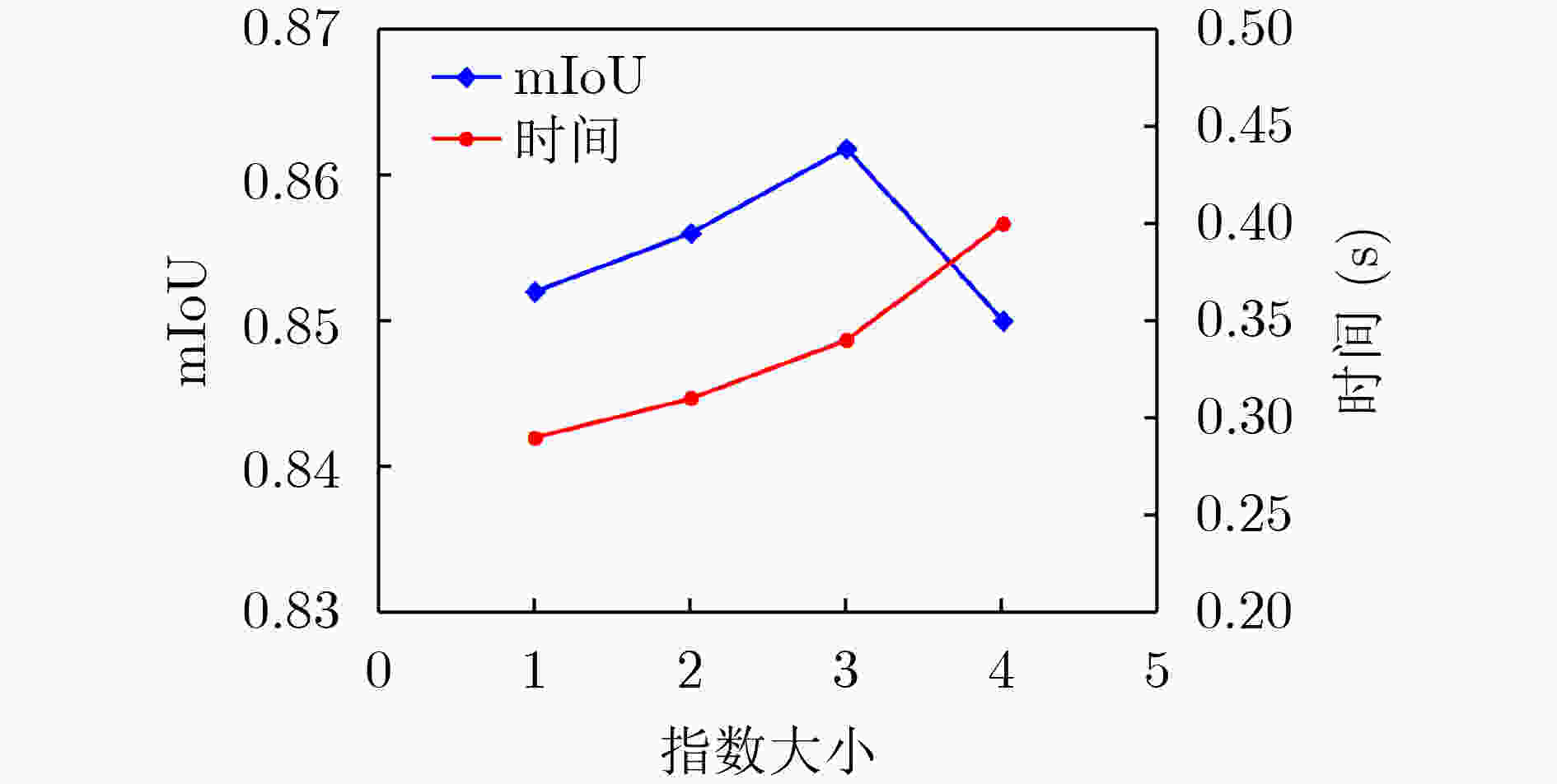



 下载:
下载:
