| [1] |
SHARMA J, CHHABRA R, CHENG A, et al. Control of self-Assembly of DNA tubules through integration of gold nanoparticles[J]. Science, 2009, 323(5910): 112–116. doi: 10.1126/science.1165831
|
| [2] |
MASTROIANNI A J, CLARIDGE S A, and ALIVISATOS A P. Pyramidal and chiral groupings of gold nanocrystals assembled using DNA scaffolds[J]. Journal of the American Chemical Society, 2009, 131(24): 8455–8459. doi: 10.1021/ja808570g
|
| [3] |
LI Bangrui, TANG Hao, YU Ruqin, et al. Single-nanoparticle ICPMS DNA assay based on hybridization-chain-reaction-mediated spherical nucleic acid assembly[J]. Analytical Chemistry, 2020, 92(3): 2379–2382. doi: 10.1021/acs.analchem.9b05741
|
| [4] |
LAN Xiang, ZHOU Xu, MCCARTHY L A, et al. DNA-enabled chiral gold nanoparticle-chromophore hybrid structure with resonant plasmon-exciton coupling gives unusual and strong circular dichroism[J]. Journal of the American Chemical Society, 2019, 141(49): 19336–19341. doi: 10.1021/jacs.9b08797
|
| [5] |
JOHNSON J A, DEHANKAR A, WINTER J O, et al. Reciprocal control of hierarchical DNA origami-nanoparticle assemblies[J]. Nano Letters, 2019, 19(12): 8469–8475. doi: 10.1021/acs.nanolett.9b02786
|
| [6] |
ZORNBERG L Z, GABRYS P A, and MACFARLANE R J. Optical processing of DNA-programmed nanoparticle superlattices[J]. Nano Letters, 2019, 19(11): 8074–8081. doi: 10.1021/acs.nanolett.9b03258
|
| [7] |
MYERS B D, PALACIOS E, MYERS D I, et al. Stimuli-responsive DNA-linked nanoparticle arrays as programmable surfaces[J]. Nano Letters, 2019, 19(7): 4535–4542. doi: 10.1021/acs.nanolett.9b01340
|
| [8] |
SHEKHIREV M, SUTTER E, and SUTTER P. In situ atomic force microscopy of the reconfiguration of on-surface self-assembled DNA-nanoparticle superlattices[J]. Advanced Functional Materials, 2019, 29(10): 1806924. doi: 10.1002/adfm.201806924
|
| [9] |
FU Wei, YOU Chao, MA Lu, et al. Enhanced efficacy of temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma[J]. ACS Applied Materials & Interfaces, 2019, 11(43): 39525–39533. doi: 10.1021/acsami.9b13829
|
| [10] |
DONG Yafei, WANG Yanchai, MA Jingjing, et al. The application of DNA nanoparticle conjugates on the graph’s connectivity problem[J]. Advances in Intelligent Systems and Computing, 2013, 212: 257–265. doi: 10.1007/978-3-642-37502-6_32
|
| [11] |
ZHANG Cheng, MA Jingjing, YANG Jing, et al. Nanoparticle aggregation logic computing controlled by DNA branch migration[J]. Applied Physics Letter, 2013, 103(9): 093106. doi: 10.1063/1.4819840
|
| [12] |
殷志祥, 唐震, 张强, 等. 基于DNA折纸基底的与非门计算模型[J]. 电子与信息学报, 2020, 42(6): 1355–1364. doi: 10.11999/JEIT190825YIN Zhixiang, TANG Zhen, ZHANG Qiang, et al. NAND gate computational model based on the DNA origami template[J]. Journal of Electronics &Information Technology, 2020, 42(6): 1355–1364. doi: 10.11999/JEIT190825
|
| [13] |
MIRKIN C A, LETSINGER R L, MUCIC R C, et al. A DNA-based method for rationally assembling nanoparticles into macroscopic materials[J]. Nature, 1996, 382(6592): 607–609. doi: 10.1038/382607a0
|
| [14] |
TATON T A, MIRKIN C A, and LETSINGER R L. Scanometric DNA array detection with nanoparticle probes[J]. Science, 2000, 289(5485): 1757–1760. doi: 10.1126/science.289.5485.1757
|
| [15] |
WEN Yongqiang, MCLAUGHLIN C K, LO P K, et al. Stable gold nanoparticle conjugation to internal DNA positions: Facile generation of discrete gold nanoparticle−DNA assemblies[J]. Bioconjugate Chemistry, 2010, 21(8): 1413–1416. doi: 10.1021/bc100160k
|
| [16] |
SHEN Xibo, SONG Chen, WANG Jinye, et al. Rolling up gold nanoparticle-dressed DNA origami into three-dimensional plasmonic chiral nanostructures[J]. Journal of the American Chemical Society, 2012, 134(1): 146–149. doi: 10.1021/ja209861x
|
| [17] |
YAO Guangbao, LI Jiang, CHAO Jie, et al. Gold-nanoparticle-mediated jigsaw-puzzle-like assembly of supersized plasmonic DNA origami[J]. Angewandte Chemie International Edition, 2015, 54(10): 2966–2969. doi: 10.1002/anie.201410895
|
| [18] |
ZANCHET D, MICHEEL C M, PARAK W J, et al. Electrophoretic isolation of discrete Au nanocrystal/DNA conjugates[J]. Nano Letters, 2001, 1(1): 32–35. doi: 10.1021/nl005508e
|
| [19] |
ZHANG Cheng, MA Jingjing, YANG Jing, et al. Binding assistance triggering attachments of hairpin DNA onto gold nanoparticles[J]. Analytical Chemistry, 2013, 85(24): 11973–11978. doi: 10.1021/ac402908y
|





 下载:
下载:
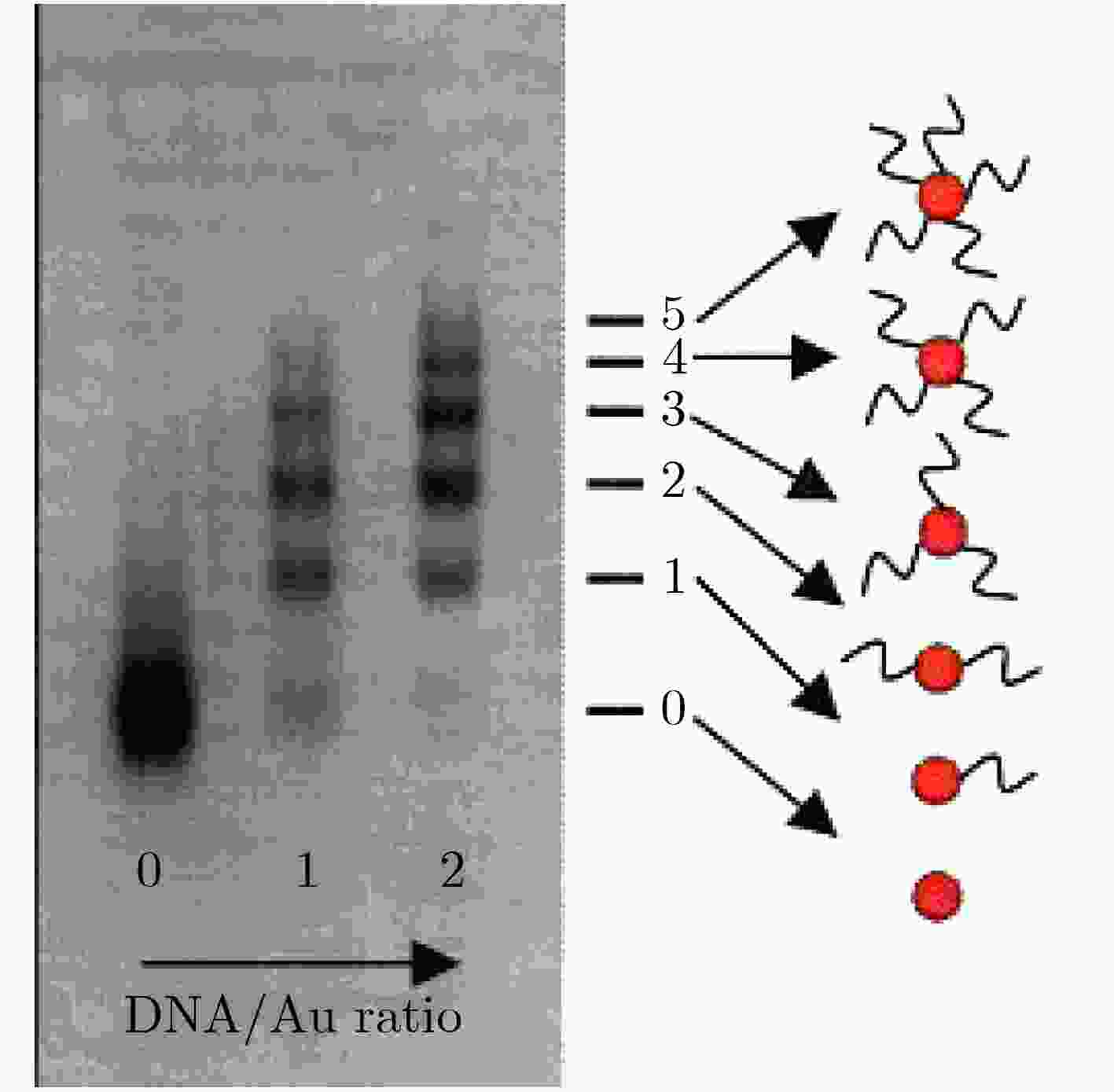
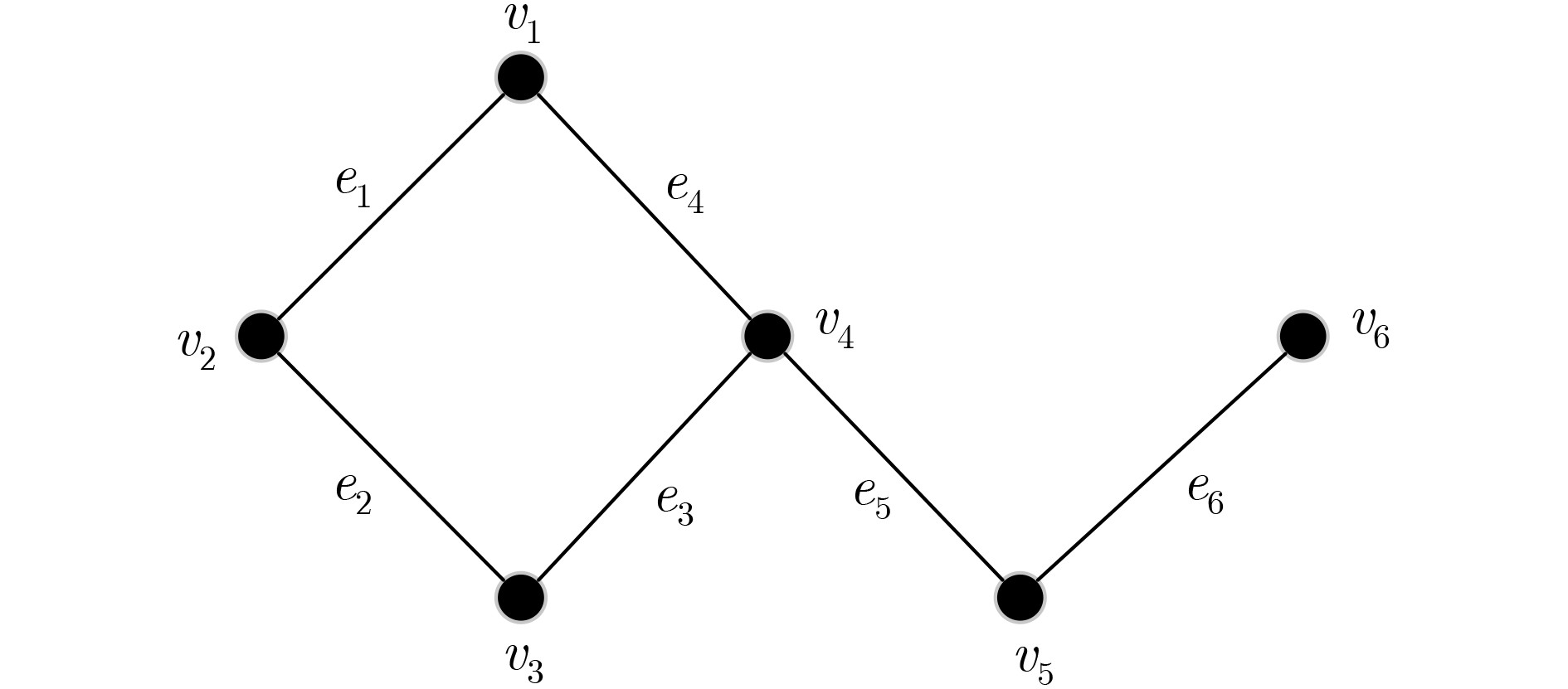


 下载:
下载:
