Study on the Differential Analysis of Alternative Splicing Based on the Median Value Jensen-Shannon Divergence
-
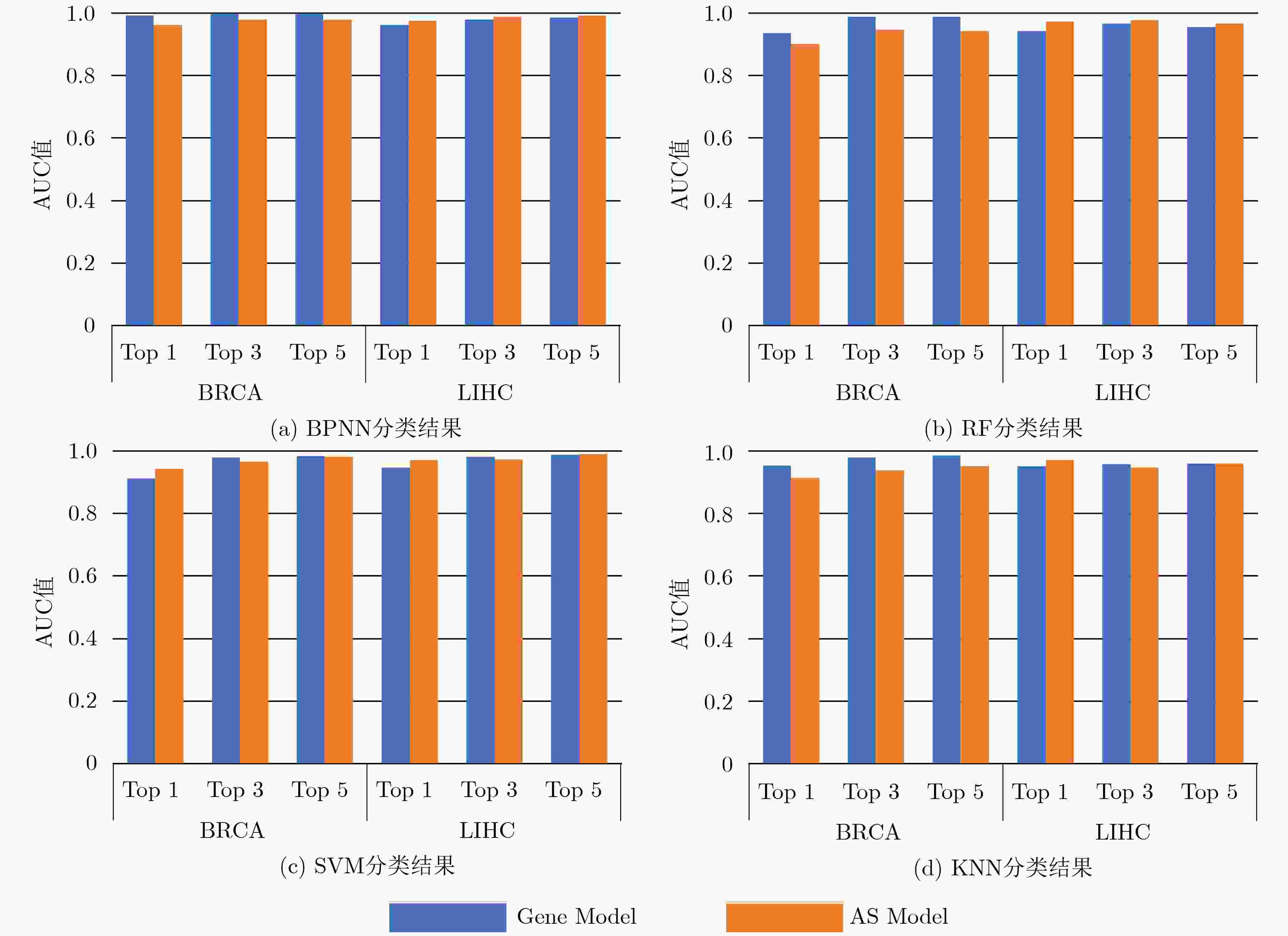
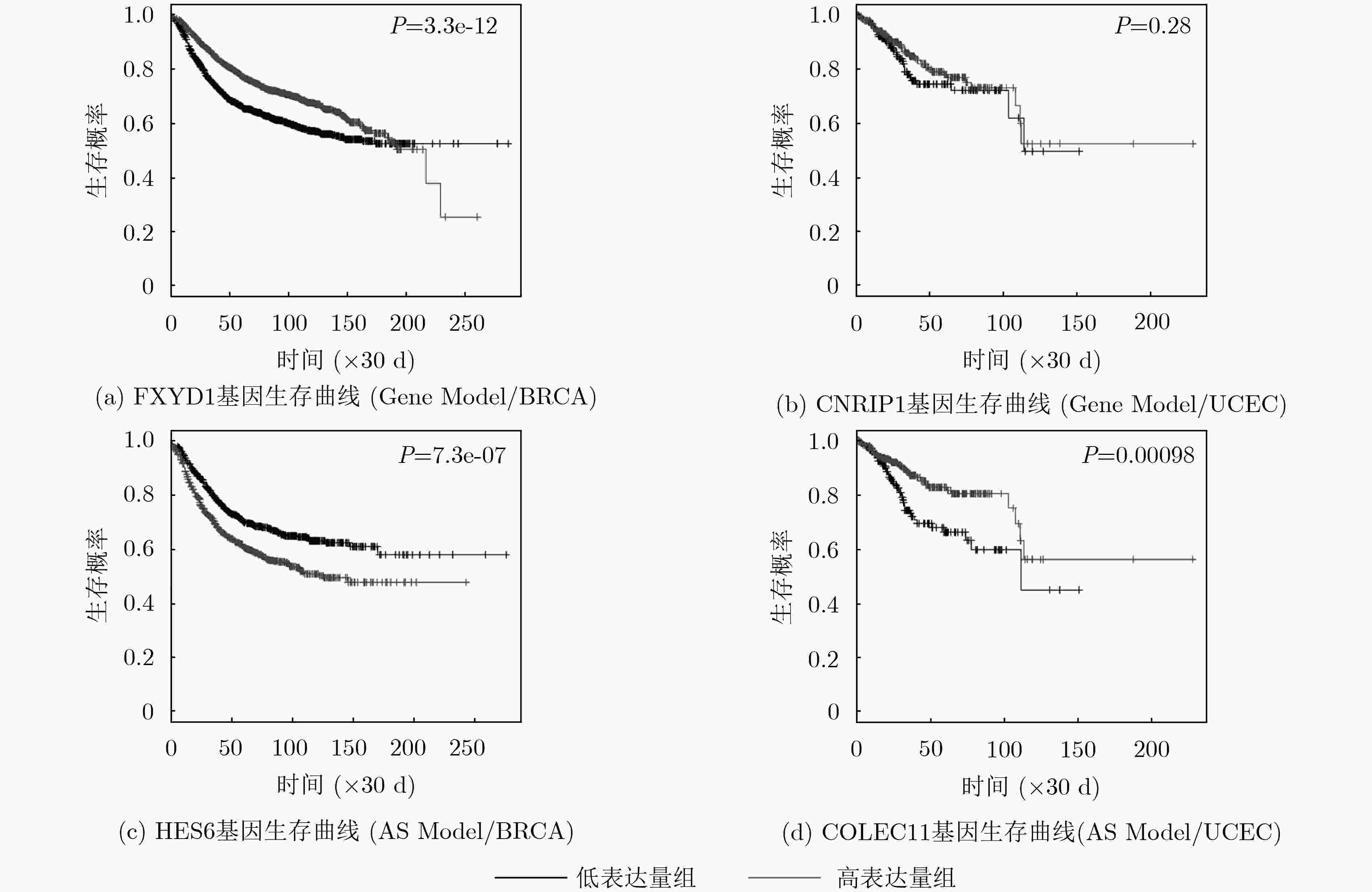
摘要: 可变剪接是一种广泛存在于生物体中造成蛋白质多样性的重要机制,它对细胞的增殖、分化、发育、凋亡等一系列重要的生物过程具有重要精细调控的作用。近年来,人们发现多种复杂疾病的产生往往伴随着剪接异构体的紊乱表达。为了研究剪接异构体在整体分布上的差异,该文提出一种基于中值的JS散度可变剪接(AS)差异分析方法。结果表明,该文的方法能够发现大量在剪接异构体整体分布上具有显著差异的基因。这些基因不仅富集在一些癌症密切相关的通路,而且也富集在一些基于可变剪接调控的信号通路、细胞分裂过程和蛋白质功能等通路。此外,与基因层次的差异分析相比,可变剪接显著差异的基因在生存分析方面也具有更好的性能。总之,该文提出基于中值的JS散度可变剪接差异分析方法,将为进一步揭示可变剪接在癌症中的机制奠定基础。Abstract: Alternative splicing is an important mechanism of protein diversity in a wide range of organisms, which plays an important role in the fine regulation of cell proliferation, differentiation, development, apoptosis and a series of important biological processes. In recent years, it is found that the occurrence of multiple complex diseases is often accompanied by the disordered expression of splicing isoforms. In order to study the difference of splicing isoforms on the whole distribution, a differential analysis method of Alternative Splicing (AS) based on the median value by Jensen-Shannon (JS) divergence is proposed in this paper. The results show the method can finds plenty of genes with significant differences in the overall distribution of splicing isoforms. These genes are not only concentrated in some cancer related pathways, but also in some signaling pathways based on alternative splicing regulation, cell division process and protein function. In addition, compared with the gene-level differential analysis, the genes with significant difference in alternative splicing also have better performance in survival analysis. In conclusion, the proposed method will lay a foundation for further revealing the mechanism of alternative splicing in cancer.
-
表 1 癌症数据集统计信息
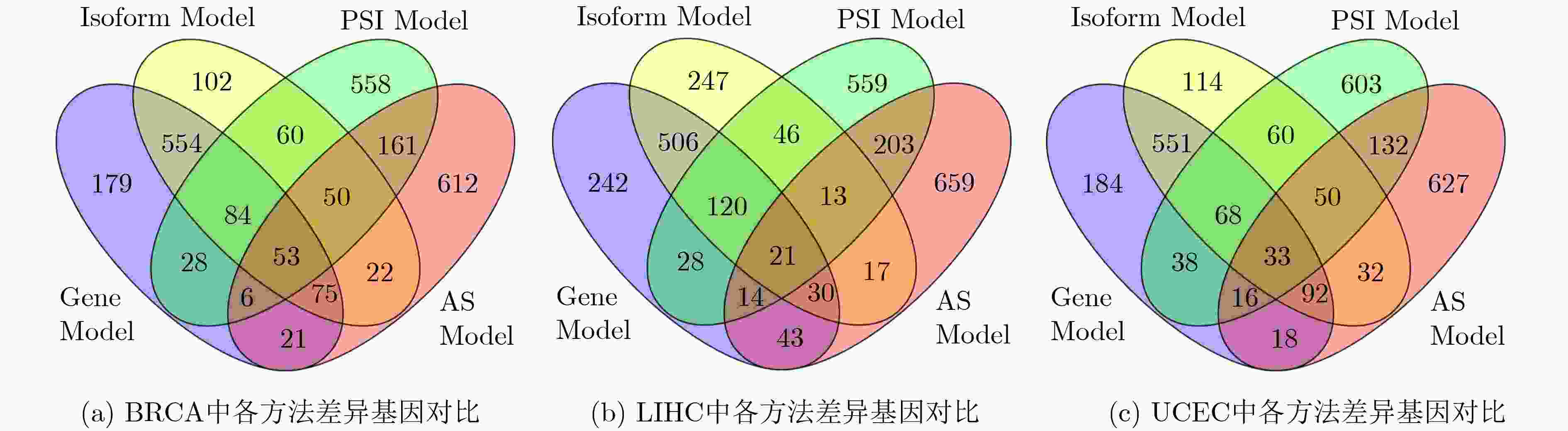
癌症 癌症样本 正常样本 基因个数 异构体个数 BRCA 1100 112 10178 33481 LIHC 373 50 8871 26234 UCEC 117 24 9765 30953 表 2 KEGG通路分析
癌症 通路(Gen Model) 通路(AS Model) Focal adhesion Cell cycle PI3K-Akt signaling pathway p53 signaling pathway Tight junction Pathways in cancer Regulation of lipolysis in adipocytes Oocyte meiosis BRCA Pathways in cancer Viral carcinogenesis Rap1 signaling pathway Adherens junction cAMP signaling pathway Purine metabolism ABC transporters PI3K-Akt signaling pathway Cell adhesion molecules (CAMs) Hippo signaling pathway Leukocyte transendothelial migration Metabolic pathways Metabolic pathways Metabolic pathways Fatty acid degradation Phagosome Protein processing in endoplasmic reticulum Fc gamma R-mediated phagocytosis Proteasome Leishmaniasis LIHC mTOR signaling pathway Homologous recombination AMPK signaling pathway Sphingolipid metabolism Valine, leucine and isoleucine degradation ECM-receptor interaction Spliceosome Cell cycle Ubiquitin mediated proteolysis Fanconi anemia pathway Insulin signaling pathway Ribosome biogenesis in eukaryotes Vascular smooth muscle contraction Osteoclast differentiation cGMP-PKG signaling pathway Cell cycle Focal adhesion Adherens junction MAPK signaling pathway Axon guidance UCEC Proteoglycans in cancer Phagosome Calcium signaling pathway Rheumatoid arthritis Platelet activation AMPK signaling pathway Adherens junction PPAR signaling pathway Oxytocin signaling pathway ECM-receptor interaction Ras signaling pathway Platelet activation -
WANG E T, SANDBERG R, LUO Shujun, et al. Alternative isoform regulation in human tissue transcriptomes[J]. Nature, 2008, 456(7221): 470–476. doi: 10.1038/nature07509 ZHOU Yujie, ZHU Guiqi, ZHANG Qingwei, et al. Survival-associated alternative messenger RNA splicing signatures in pancreatic ductal adenocarcinoma: A study based on RNA-sequencing data[J]. DNA and Cell Biology, 2019, 38(11): 1207–1222. doi: 10.1089/dna.2019.4862 XIE Zucheng, WU Huayu, DANG Yiwu, et al. Role of alternative splicing signatures in the prognosis of glioblastoma[J]. Cancer Medicine, 2019, 8(18): 7623–7636. doi: 10.1002/cam4.2666 LI Mingxue, WANG Dun, HE Jianhua, et al. Bcl-XL: A multifunctional anti-apoptotic protein[J]. Pharmacological Research, 2020, 151: 104547. doi: 10.1016/j.phrs.2019.104547 KOLE R, KRAINER A R, and ALTMAN S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides[J]. Nature Reviews Drug Discovery, 2012, 11(2): 125–140. doi: 10.1038/nrd3625 SONG Jukun, LIU Yongda, SU Jiaming, et al. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma[J]. Journal of Cellular Physiology, 2019, 234(12): 22753–22764. doi: 10.1002/jcp.28840 DOU Tonghai, XU Jiaxi, GAO Yuan, et al. Evolution of peroxisome proliferator-activated receptor gamma alternative splicing[J]. Frontiers in Bioscience (Elite Edition) , 2010, 2: 1334–1343. doi: 10.2741/e193 LI Ji, CHOI P S, CHAFFER C L, et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer[J]. eLife, 2018, 7: e37184. doi: 10.7554/eLife.37184 SHEN Shihao, PARK J W, LU Zhixiang, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(51): E5593–E5601. doi: 10.1073/pnas.1419161111 欧书华, 刘学军, 张礼. 基于KL散度的RNA-Seq数据差异异构体比例检测[J]. 计算机工程与科学, 2017, 39(1): 158–164. doi: 10.3969/j.issn.1007-130X.2017.01.022OU Shuhua, LIU Xuejun, and ZHANG Li. Differential isoform ratio detection based on KL divergence for RNA-Seq data[J]. Computer Engineering &Science, 2017, 39(1): 158–164. doi: 10.3969/j.issn.1007-130X.2017.01.022 LIU Jingwei, LI Hao, SHEN Shixuan, et al. Alternative splicing events implicated in carcinogenesis and prognosis of colorectal cancer[J]. Journal of Cancer, 2018, 9(10): 1754–1764. doi: 10.7150/jca.24569 ZONG Zhen, LI Hui, YI Chenghao, et al. Genome-wide profiling of prognostic alternative splicing signature in colorectal cancer[J]. Frontiers in Oncology, 2018, 8: 537. doi: 10.3389/fonc.2018.00537 ZHANG Zijun, PAN Zhicheng, YING Yi, et al. Deep-learning augmented RNA-seq analysis of transcript splicing[J]. Nature Methods, 2019, 16(4): 307–310. doi: 10.1038/s41592-019-0351-9 WHITFIELD M L, GEORGE L K, GRANT G D, et al. Common markers of proliferation[J]. Nature Reviews Cancer, 2006, 6(2): 99–106. doi: 10.1038/nrc1802 INOUE K and FRY E A. Aberrant splicing of the DMP1-ARF-MDM2-p53 pathway in cancer[J]. International Journal of Cancer, 2016, 139(1): 33–41. doi: 10.1002/ijc.30003 MUNDING E M, SHIUE L, KATZMAN S, et al. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing[J]. Molecular Cell, 2013, 51(3): 338–348. doi: 10.1016/j.molcel.2013.06.012 CHU Xiufeng, ZHANG Ting, WANG Jie, et al. Alternative splicing variants of human Fbx4 disturb cyclin D1 proteolysis in human cancer[J]. Biochemical and Biophysical Research Communications, 2014, 447(1): 158–164. doi: 10.1016/j.bbrc.2014.03.129 BAILEY M H, TOKHEIM C, PORTA-PARDO E, et al. Comprehensive characterization of cancer driver genes and mutations[J]. Cell, 2018, 173(2): 371–385. e18. doi: 10.1016/j.cell.2018.02.060 曾勇, 舒欢, 胡江平, 等. 基于BP神经网络的自适应伪最近邻分类[J]. 电子与信息学报, 2016, 38(11): 2774–2779. doi: 10.11999/JEIT160133ZENG Yong, SHU Huan, HU Jiangping, et al. Adaptive pseudo nearest neighbor classification based on BP neural network[J]. Journal of Electronics &Information Technology, 2016, 38(11): 2774–2779. doi: 10.11999/JEIT160133 陈素根, 吴小俊. 基于特征值分解的中心支持向量机算法[J]. 电子与信息学报, 2016, 38(3): 557–564. doi: 10.11999/JEIT150693CHEN Sugen and WU Xiaojun. Eigenvalue proximal support vector machine algorithm based on eigenvalue decoposition[J]. Journal of Electronics &Information Technology, 2016, 38(3): 557–564. doi: 10.11999/JEIT150693 ZHANG Yangjun, YAN Libin, ZENG Jin, et al. Pan-cancer analysis of clinical relevance of alternative splicing events in 31 human cancers[J]. Oncogene, 2019, 38(40): 6678–6695. doi: 10.1038/s41388-019-0910-7 -





 下载:
下载:



 下载:
下载:
