Design of Three-cascade Combinatorial Molecular Logic Circuit Based on DNA Strand Displacement
-
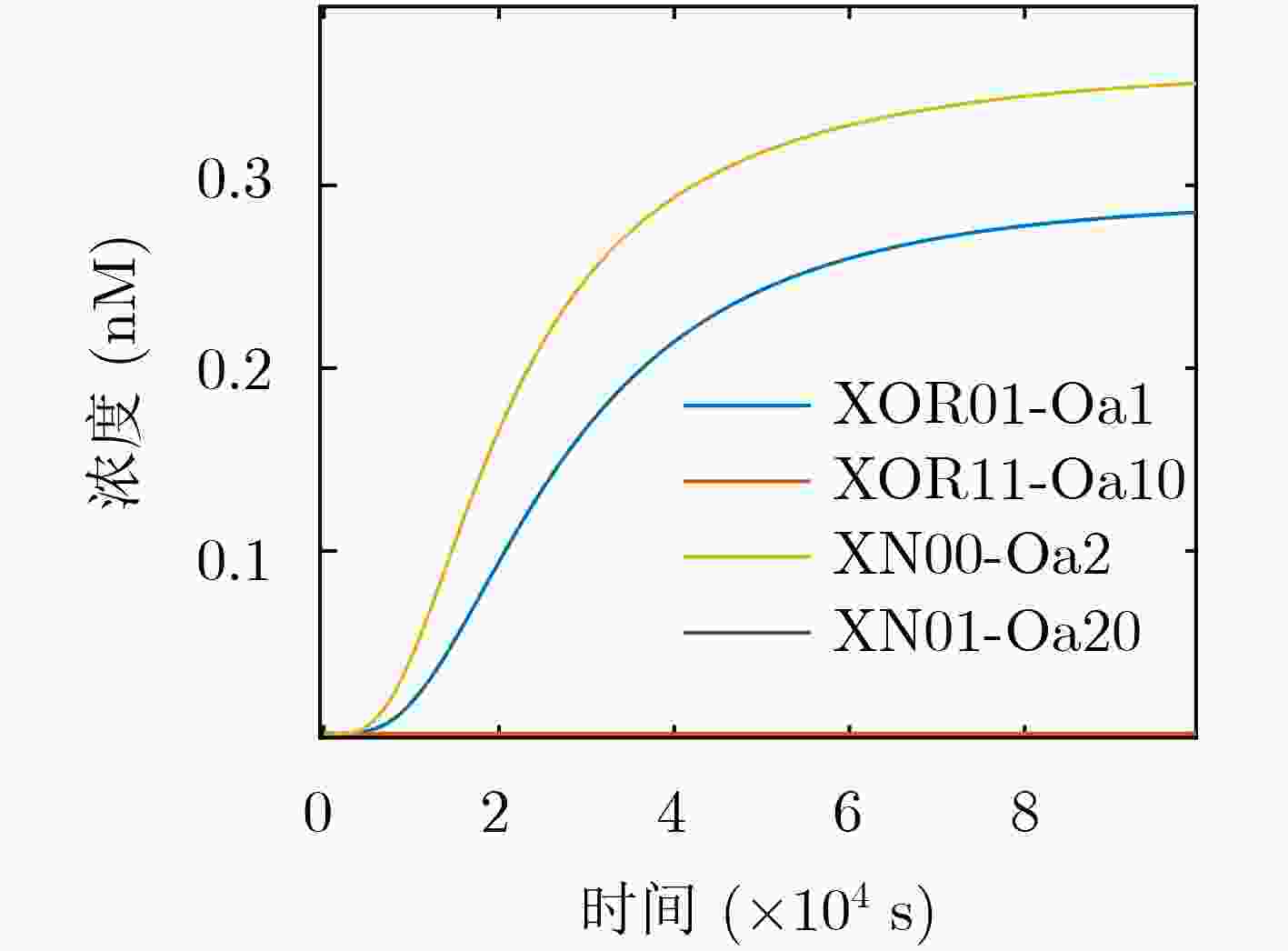
摘要: DNA计算研究内容繁多复杂,DNA复杂逻辑电路的搭建属于DNA计算的一个重要研究分支,其中逻辑门的构建属于DNA复杂逻辑电路搭建的基础研究,设计出更为简单的逻辑门可以为研究者搭建复杂电路提供参考,节省基础研究的宝贵时间。针对上述问题,该文利用使能控制端思想,采用DNA链置换技术,设计了与或、与非或非和异或同或3种DNA组合逻辑门。结果显示,设计的3种组合逻辑门可实现6种逻辑运算功能,并利用所构建的组合逻辑门成功构造了多级联组合分子逻辑电路,为DNA计算提供了更多的解决方案,促进了DNA计算机的发展。Abstract: The research content of DNA computing is various and complex. The construction of DNA complex logic circuit belongs to an important research branch of DNA computing, in which the construction of logic gate belongs to the basic research of DNA complex logic circuit construction. The design of a simpler logic gate is used to provide a reference for researchers to build complex circuits and save valuable time for basic research. In order to solve the above problems, the idea of enable control end and DNA strand displacement technique are used to design three kinds of DNA combinatorial logic gates: AND-OR gate, NAND-NOR gate and XOR-XNOR gate. The results show that the three kinds of combinatorial logic gates can realize six kinds of logic operation functions, and the multi-stage combinatorial molecular logic circuits are successfully constructed by using the combinatorial logic gates, which provides more solutions for DNA calculation. It promotes the development of DNA computer.
-
Key words:
- DNA computing /
- DNA strand displacement /
- DNA combinatorial logic gates
-
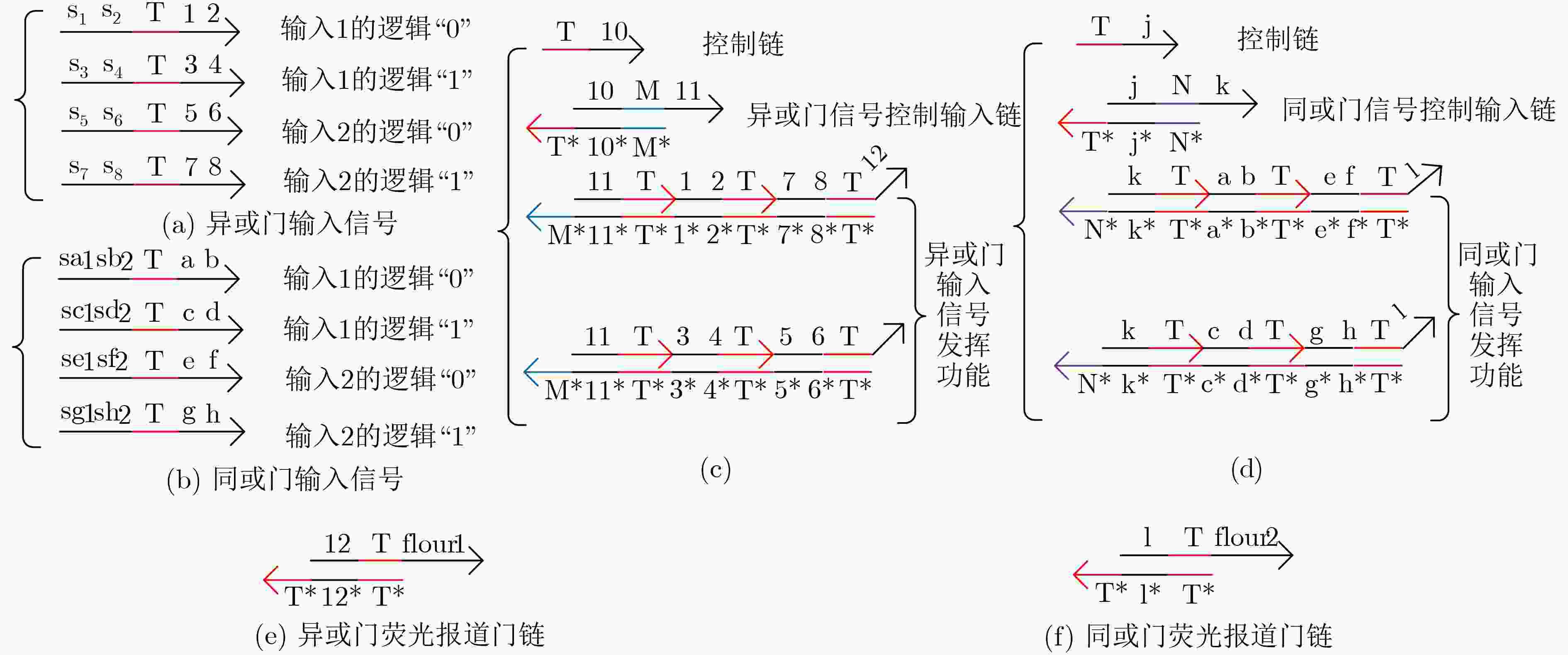
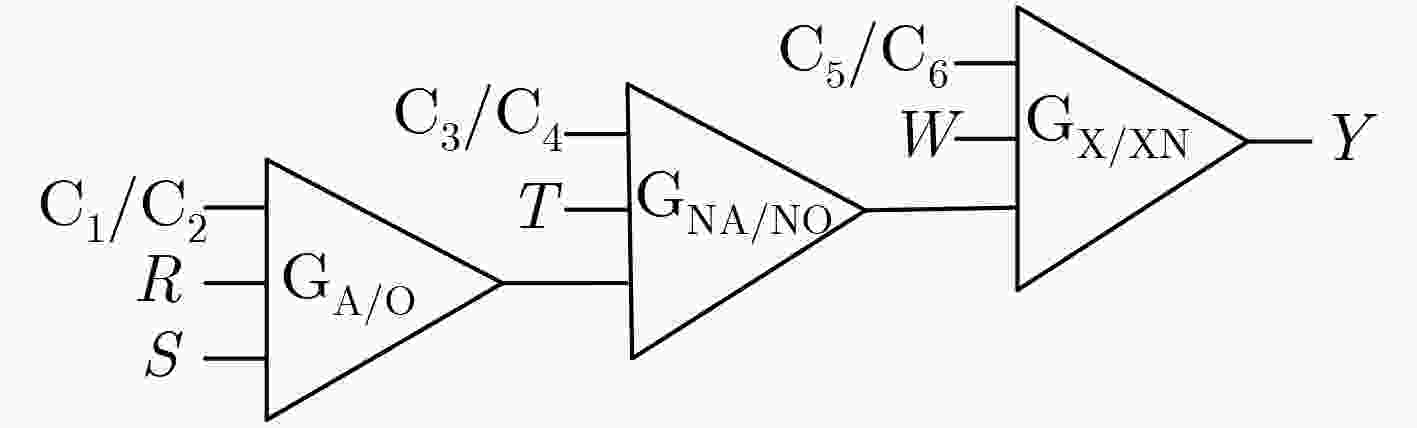
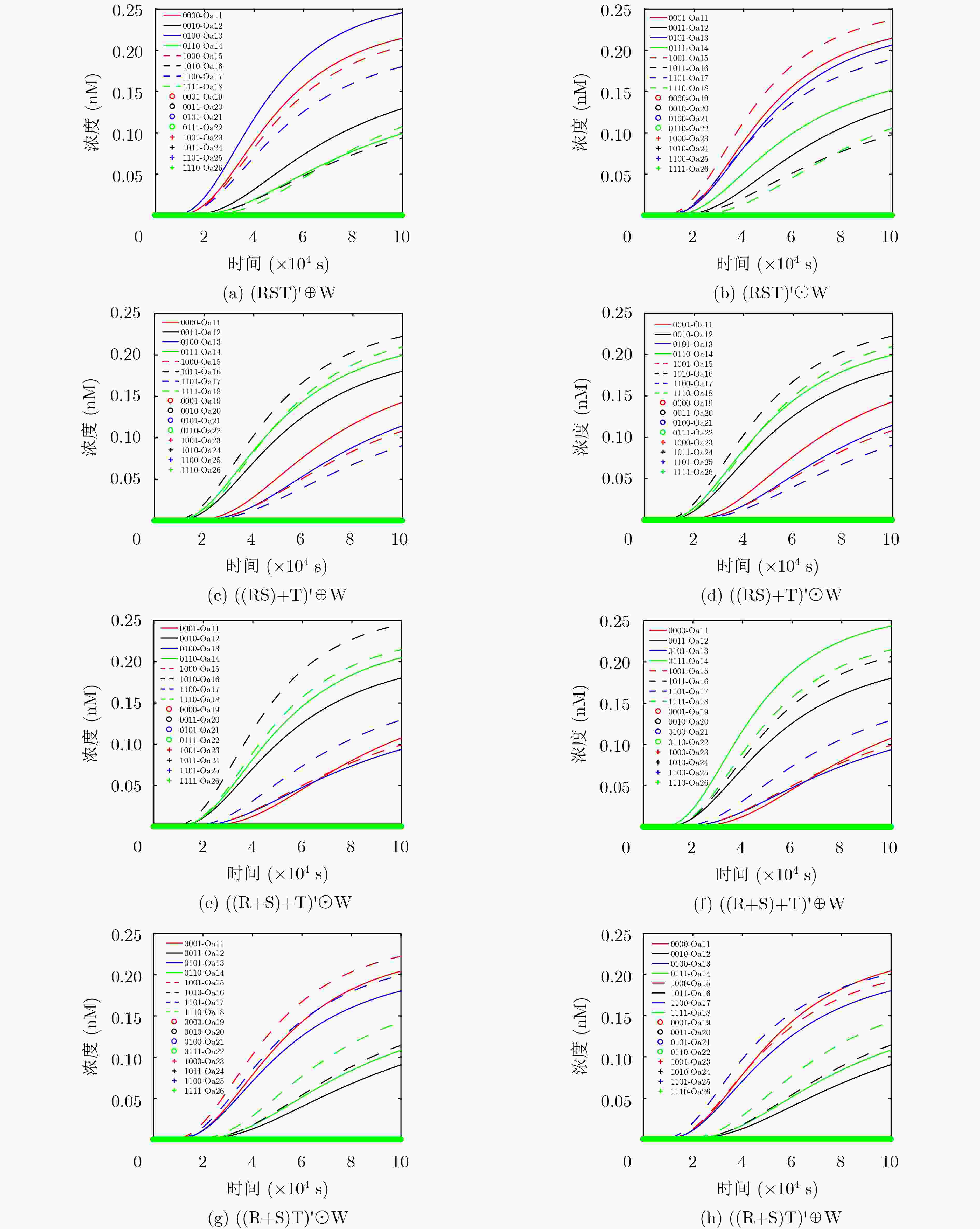
表 1 4输入3级联组合逻辑电路真值表
序号 C1/C2 C3/C4 C5/C6 Y 序号 C1/C2 C3/C4 C5/C6 Y 1 ON/OFF ON/OFF ON/OFF (ABC)'⊕D 6 OFF/ON OFF/ON ON/OFF ((A+B)+C)'⊕D 2 ON/OFF ON/OFF OFF/ON (ABC)'⊙D 7 OFF/ON ON/OFF OFF/ON ((A+B)C)'⊙D 3 ON/OFF OFF/ON ON/OFF ((AB)+C)'⊕D 8 OFF/ON ON/OFF ON/OFF ((A+B)C)'⊕D 4 ON/OFF OFF/ON OFF/ON ((AB)+C)'⊙D 9 OFF/OFF OFF/OFF OFF/OFF OFF 5 OFF/ON OFF/ON OFF/ON ((A+B)+C)'⊙D 10 ON/ON ON/ON ON/ON ON -
殷志祥, 唐震, 张强, 等. 基于DNA折纸基底的与非门计算模型[J]. 电子与信息学报, 2020, 42(6): 1355–1364. doi: 10.11999/JEIT190825YIN Zhixiang, TANG Zhen, ZHANG Qiang, et al. NAND gate computational model based on the DNA origami template[J]. Journal of Electronics &Information Technology, 2020, 42(6): 1355–1364. doi: 10.11999/JEIT190825 梁静, 李红菊, 赵凤, 等. 一种构造GC常重量DNA码的方法[J]. 电子与信息学报, 2019, 41(10): 2423–2427. doi: 10.11999/JEIT190070LIANG Jing, LI Hongju, ZHAO Feng, et al. A method for constructing GC constant weight DNA codes[J]. Journal of Electronics &Information Technology, 2019, 41(10): 2423–2427. doi: 10.11999/JEIT190070 ADLEMAN L M. Molecular computation of solutions to combinatorial problems[J]. Science, 1994, 266(5187): 1021–1024. doi: 10.1126/science.7973651 LAKIN M R, YOUSSEF S, POLO F, et al. Visual DSD: A design and analysis tool for DNA strand displacement systems[J]. Bioinformatics, 2011, 27(22): 3211–3213. doi: 10.1093/bioinformatics/btr543 ZHU Jinbo, ZHANG Libing, DONG Shaojun, et al. Four-way junction-driven DNA strand displacement and its application in building majority logic circuit[J]. ACS Nano, 2013, 7(11): 10211–10217. doi: 10.1021/nn4044854 KONG Jinglin, ZHU Jinbo, CHEN Kaikai, et al. Specific biosensing using DNA aptamers and nanopores[J]. Advanced Functional Materials, 2019, 29(3): 180755. doi: 10.1002/adfm.201807555 CUI Yunxi, FENG Xuenan, WANG Yaxin, et al. An integrated-molecular-beacon based multiple exponential strand displacement amplification strategy for ultrasensitive detection of DNA methyltransferase activity[J]. Chemical Science, 2019, 10(8): 2290–2297. doi: 10.1039/C8SC05102J LI Hua, LIU Jin, and GU Hongzhou. Targeting nucleolin to obstruct vasculature feeding with an intelligent DNA nanorobot[J]. Journal of Cellular and Molecular Medicine, 2019, 23(3): 2248–2250. doi: 10.1111/jcmm.14127 TIKHOMIROV G, PETERSEN P, and QIAN Lulu. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns[J]. Nature, 2017, 552(7683): 67–71. doi: 10.1038/nature24655 KIELAR C, REDDAVIDE F V, TUBBENHAUER S, et al. Pharmacophore nanoarrays on DNA origami substrates as a single-molecule assay for fragment-based drug discovery[J]. Angewandte Chemie, 2018, 130(45): 15089–15093. doi: 10.1002/ange.201806778 TASCIOTTI E. Smart cancer therapy with DNA origami[J]. Nature Biotechnology, 2018, 36(3): 234–235. doi: 10.1038/nbt.4095 CORDEIRO M, OTRELO-CARDOSO A R, SVERGUN D I, et al. Optical and structural characterization of a chronic myeloid leukemia DNA biosensor[J]. ACS Chemical Biology, 2018, 13(5): 1235–1242. doi: 10.1021/acschembio.8b00029 QIAN Lulu and WINFREE E. A simple DNA gate motif for synthesizing large-scale circuits[J]. Journal of the Royal Society Interface, 2011, 8(62): 1281–1297. doi: 10.1098/rsif.2010.0729 WUNSCH B H, KIM S C, GIFFORD S M, et al. Gel-on-a-chip: Continuous, velocity-dependent DNA separation using nanoscale lateral displacement[J]. Lab on a Chip, 2019, 19(9): 1567–1578. doi: 10.1039/C8LC01408F 王春华, 蔺海荣, 孙晶如, 等. 基于忆阻器的混沌、存储器及神经网络电路研究进展[J]. 电子与信息学报, 2020, 42(4): 795–810. doi: 10.11999/JEIT190821WANG Chunhua, LIN Hairong, SUN Jingru, et al. Research Progress on Chaos, Memory and Neural Network Circuits Based on Memristor[J]. Journal of Electronics and Information Technology, 2020, 42(4): 795–810. doi: 10.11999/JEIT190821 HE Jinglin, ZHANG Yang, YANG Chan, et al. Hybridization chain reaction based DNAzyme fluorescent sensor for L-histidine assay[J]. Analytical Methods, 2019, 11(16): 2204–2210. doi: 10.1039/C9AY00526A LIU Na, XU Kai, LIU Liquan, et al. A star-shaped DNA probe based on strand displacement for universal and multiplexed fluorometric detection of genetic variations[J]. Microchimica Acta, 2018, 185(9): 413. doi: 10.1007/s00604-018-2941-0 ZOU Chengye, WEI Xiaopeng, ZHANG Qiang, et al. Four-analog computation based on DNA strand displacement[J]. ACS Omega, 2017, 2(8): 4143–4160. doi: 10.1021/acsomega.7b00572 SUN Junwei, LI Xing, CUI Guangzhao, et al. One-bit half adder-half subtractor logical operation based on the DNA strand displacement[J]. Journal of Nanoelectronics and Optoelectronics, 2017, 12(4): 375–380. doi: 10.1166/jno.2017.2027 LI Wei, YANG Yang, YAN Hao, et al. Three-input majority logic gate and multiple input logic circuit based on DNA strand displacement[J]. Nano Letters, 2013, 13(6): 2980–2988. doi: 10.1021/nl4016107 张成, 马丽娜, 董亚非, 等. 自组装DNA链置换分子逻辑计算模型[J]. 科学通报, 2012, 57(31): 2909–2915. doi: 10.1360/csb2012-57-31-2909ZHANG Cheng, MA Lina, DONG Yafei, et al. Molecular logic computing model based on DNA self-assembly strand branch migration[J]. Chinese Science Bulletin, 2012, 57(31): 2909–2915. doi: 10.1360/csb2012-57-31-2909 -





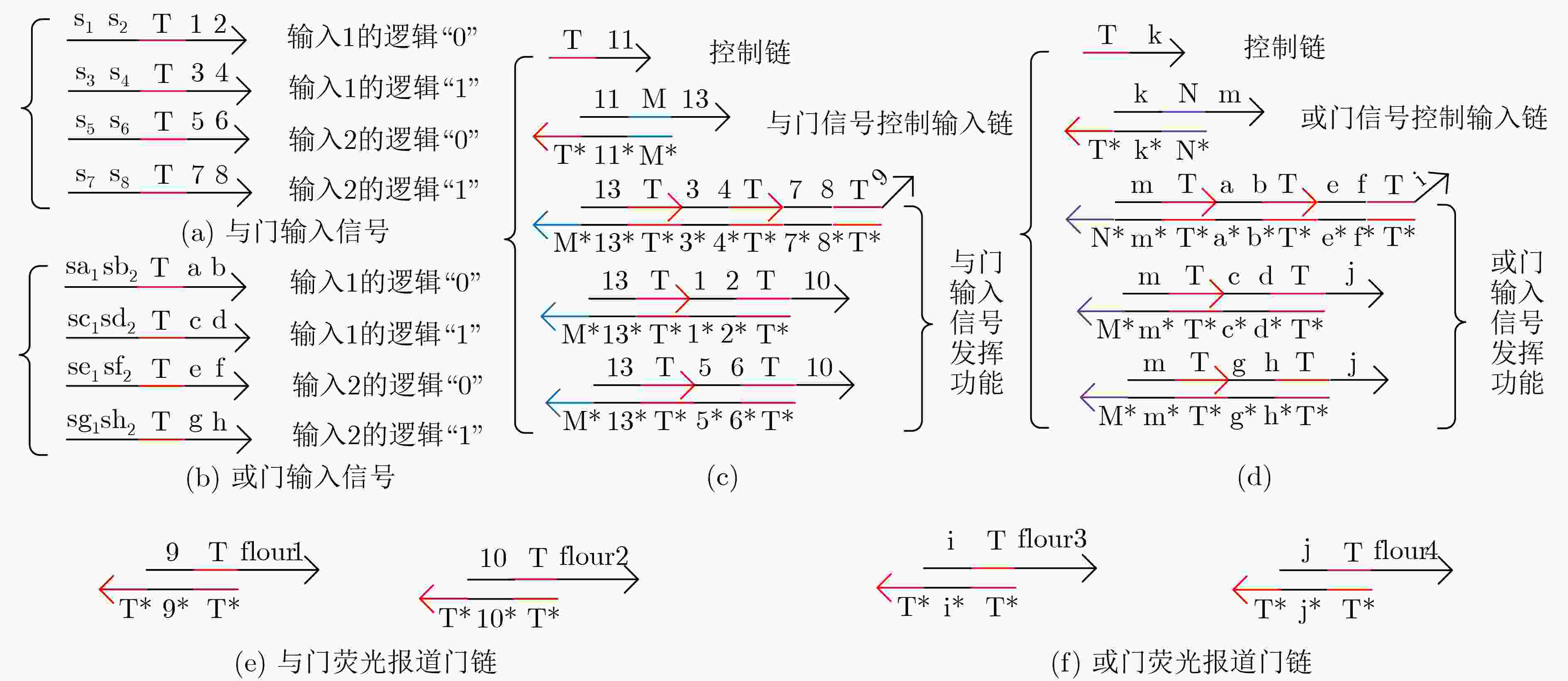
 下载:
下载:
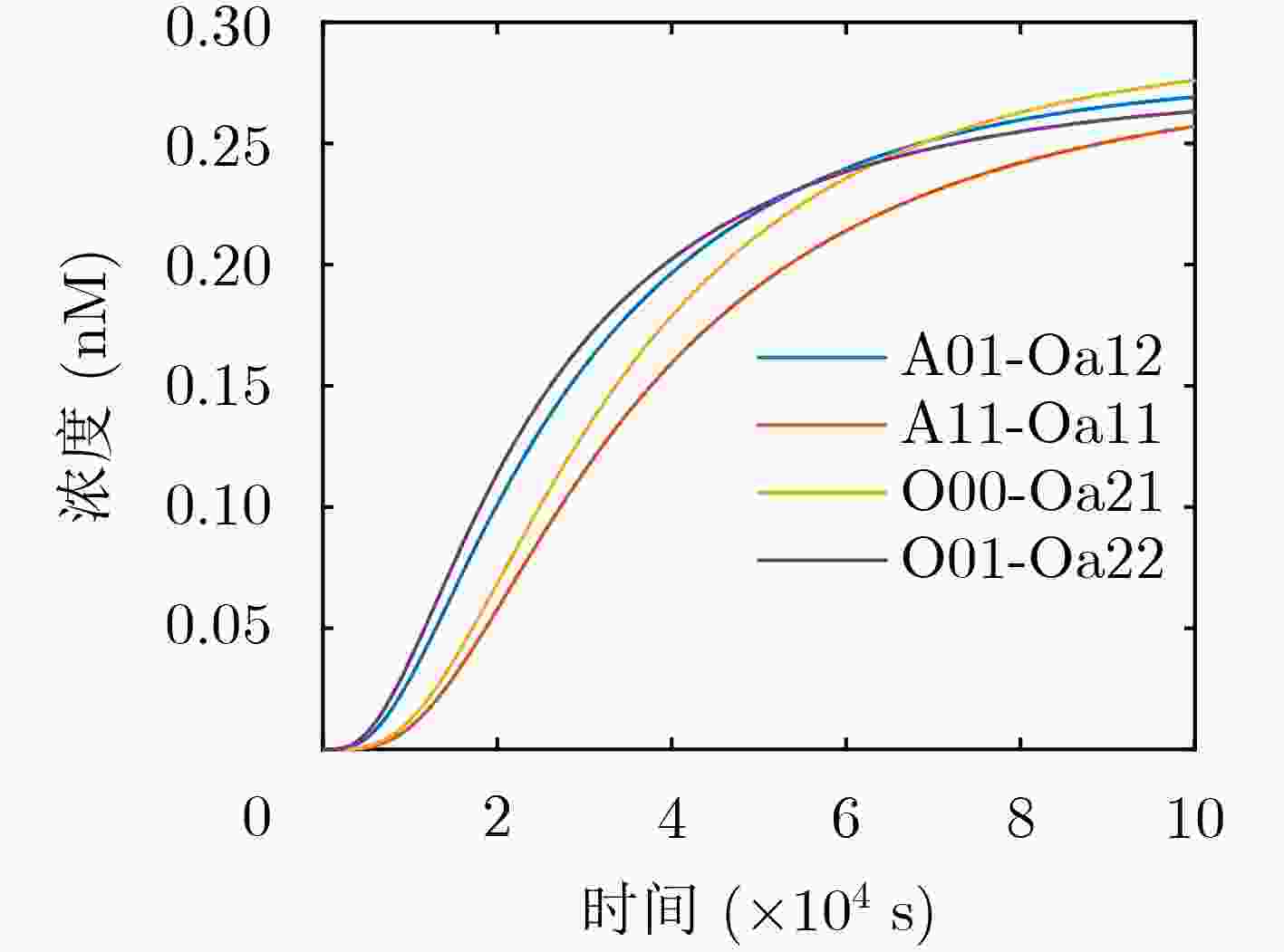
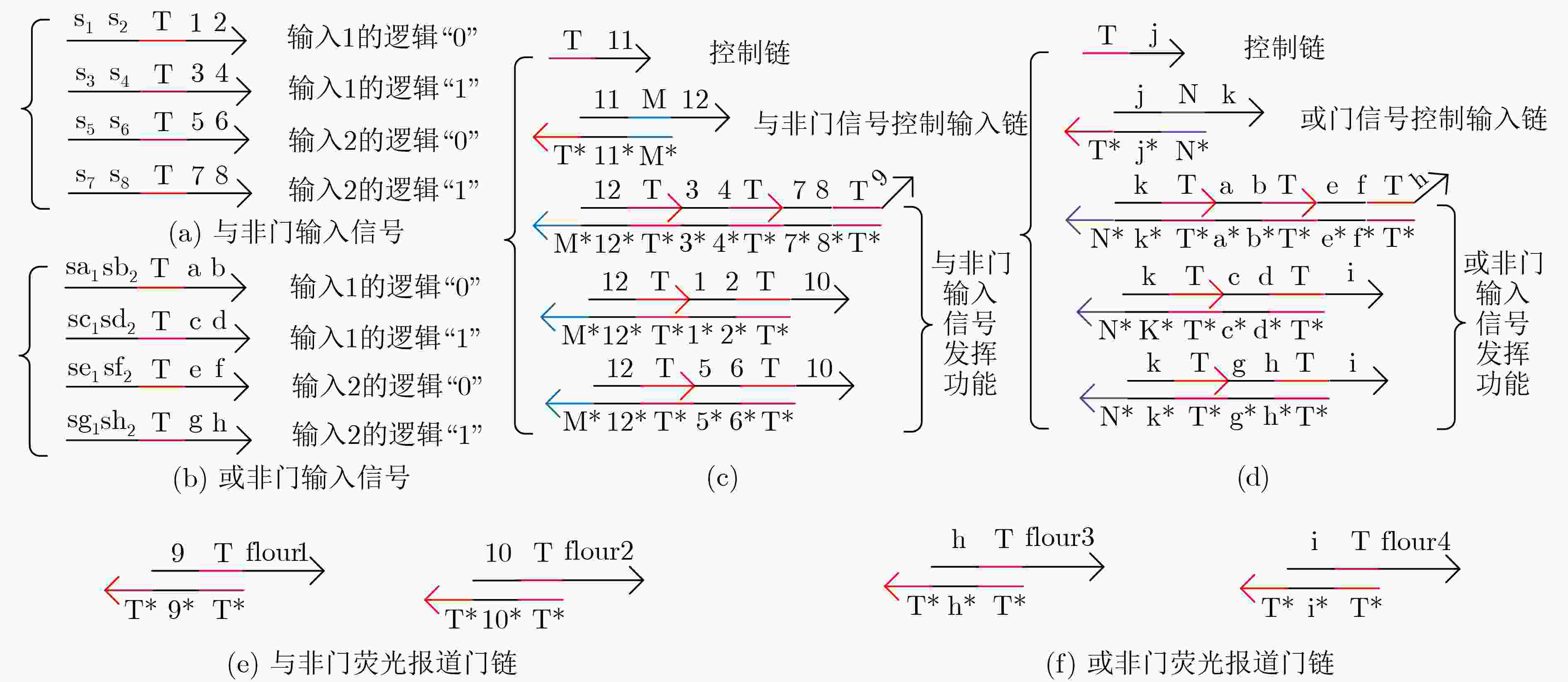
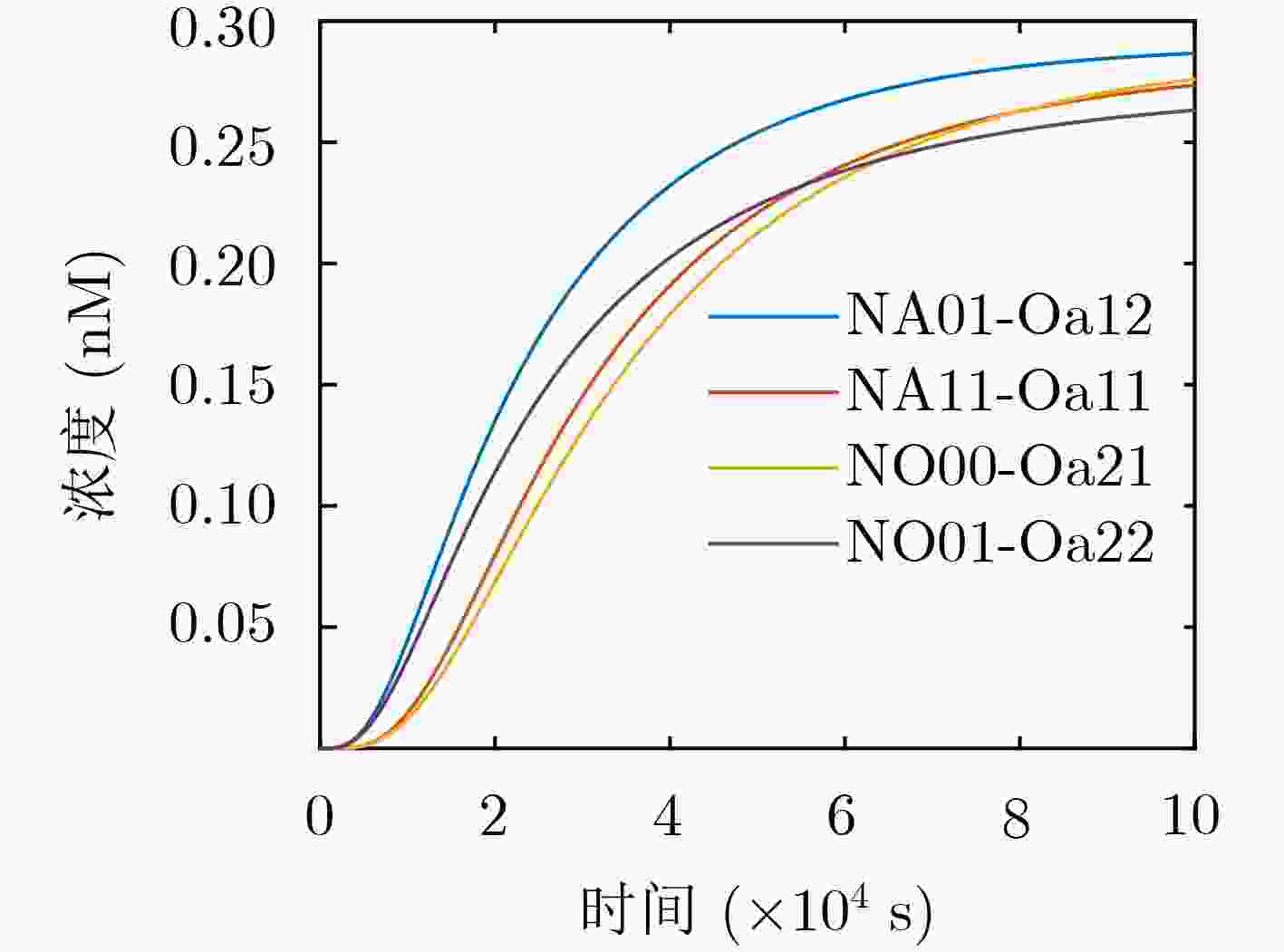


 下载:
下载:
