Basic Logic Gates Design Based on DNA and Restriction Endonuclease
-
摘要: 由于DNA分子具有特异性、高并行性、微小性等天然特性,在信息处理过程中展现出了强大的并行计算能力和数据存储能力。该文研究将具有特异性识别功能的限制性核酸内切酶引入DNA链置换反应中,作为DNA电路的输入,通过控制立足点的生成和移除设计了是门、非门和与门3种基本逻辑门。采用Visual DSD对逻辑模型进行模拟仿真,并通过凝胶电泳实验验证设计。与以往的分子逻辑门比较,该设计反应迅速,操作简便,具有良好的扩展性,为大规模电路的设计提供了可能性。Abstract: Due to the natural characteristics of specificity, high parallelism and miniaturization of DNA molecules, it exhibits strong parallel computing power and data storage capability in information processing. In this study, restriction endonuclease with specific recognition function are introduced into DNA strand displacement as the input of the DNA circuit. The YES gate, NOT gate and AND gate are designed by controlling the generation and removal of the toehold. The logic model is simulated by Visual DSD, and the design is verified by PolyacrylAmide Gel Electrophoresis(PAGE) experiments. Compared with previous molecular logic gates, this design has a quick response, simple operation, and good scalability, which provides the possibility for the design of large-scale circuits.
-
表 1 实验所需寡核苷酸序列
DNA链 序列(5’ to 3’) 所涉及逻辑门 起作用的限制酶 A TTTTTTTTTTTGATCCGTTCCTTGCAGTTGCTGAGGTGGCCAT 非门 / B TTATGGCCACCTCAGCAACTGCAAGGAACGGATCA 非门 Nt.BbvCI C TGATCCGTTCCTTGCAGTTGCTGAGGT 非门 / P GGCTGCGAGACTCGGTTTTCCGAGTCTCGCAGCCTCAGCAGTTGGATACATCTCAAGC(其中TTTT为环形结构) 与门 Nt.BsmAI X TTTTTTTCAGCCTCAGCAGTTGGATACATCTCAAGCTTTTTTTTTTTTTTT 是门、与门 Nt.BbvCI Y GCTTGAGATGTATCCAACTGCTGAGGCTG 是门、与门 / Z CAGCCTCAGCAGTTGGATACATCTCAAGC 是门 / -
WALDROP M M. The chips are down for Moore’s law[J]. Nature, 2016, 530(7589): 144–147. doi: 10.1038/530144a 梁静, 李红菊, 赵凤, 等. 一种构造GC常重量DNA码的方法[J]. 电子与信息学报, 2019, 41(10): 2423–2427. doi: 10.11999/JEIT190070LIANG Jing, LI Hongju, ZHAO Feng, et al. A method for constructing GC constant weight DNA codes[J]. Journal of Electronics &Information Technology, 2019, 41(10): 2423–2427. doi: 10.11999/JEIT190070 李歆昊, 张旻. 基于人工免疫的链路层协议帧同步字识别[J]. 电子与信息学报, 2017, 39(3): 561–567. doi: 10.11999/JEIT160476LI Xinhao and ZHANG Min. Frame synchronization word identification of link layer protocol based on artificial immune[J]. Journal of Electronics &Information Technology, 2017, 39(3): 561–567. doi: 10.11999/JEIT160476 BONNET J, YIN P, ORTIZ M E, et al. Amplifying genetic logic gates[J]. Science, 2013, 340(6132): 599–603. doi: 10.1126/science.1232758 CHATTERJEE G, DALCHAU N, MUSCAT R A, et al. A spatially localized architecture for fast and modular DNA computing[J]. Nature Nanotechnology, 2017, 12(9): 920–927. doi: 10.1038/nnano.2017.127 WEINBERG B H, PHAM N T H, CARABALLO L D, et al. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells[J]. Nature Biotechnology, 2017, 35(5): 453–462. doi: 10.1038/nbt.3805 GARG S, SHAH S, BUI H, et al. Renewable time-responsive DNA circuits[J]. Small, 2018, 14(33): 1801470. doi: 10.1002/smll.201801470 张成, 杨静, 王淑栋. DNA计算中荧光技术的应用及其发展[J]. 计算机学报, 2009, 32(12): 2300–2310. doi: 10.3724/SP.J.1016.2009.02300ZHANG Cheng, YANG Jing, and WANG Shudong. Development and application of fluorescence technology in DNA computing[J]. Chinese Journal of Computers, 2009, 32(12): 2300–2310. doi: 10.3724/SP.J.1016.2009.02300 张川, 钟志伟, 庄雨辰, 等. 面向组合逻辑的DNA计算[J]. 中国科学: 信息科学, 2019, 49(7): 819–837. doi: 10.1360/N112019-00007ZHANG Chuan, ZHONG Zhiwei, ZHUANG Yuchen, et al. DNA computing for combinational logic[J]. Scientia Sinica Informationis, 2019, 49(7): 819–837. doi: 10.1360/N112019-00007 YURKE B, TURBERFIELD A J, MILLS JR A P, et al. A DNA-fuelled molecular machine made of DNA[J]. Nature, 2000, 406(6796): 605–608. doi: 10.1038/35020524 LI Wei, ZHANG Fei, YAN Hao, et al. DNA based arithmetic function: A half adder based on DNA strand displacement[J]. Nanoscale, 2016, 8(6): 3775–3784. doi: 10.1039/C5NR08497K ZHANG Tianchi, SHANG Chunli, DUAN Ruixue, et al. Polar organic solvents accelerate the rate of DNA strand replacement reaction[J]. The Analyst, 2015, 140(6): 2023–2028. doi: 10.1039/C4AN02302A YANG Xiaolong, TANG Yanan, TRAYNOR S M, et al. Regulation of DNA strand displacement using an allosteric DNA toehold[J]. Journal of the American Chemical Society, 2016, 138(42): 14076–14082. doi: 10.1021/jacs.6b08794 张文逸, 殷志祥. 基于DNA链置换的分子逻辑门计算模型[J]. 安徽理工大学学报: 自然科学版, 2015, 35(1): 7–10, 34. doi: 10.3969/j.issn.1672-1098.2015.01.002ZHANG Wenyi and YIN Zhixiang. Calculation model of molecular logic gates based on DNA strand displacement[J]. Journal of Anhui University of Science and Technology:Natural Science, 2015, 35(1): 7–10, 34. doi: 10.3969/j.issn.1672-1098.2015.01.002 PAN Linqiang, WANG Zhiyu, LI Yifan, et al. Nicking enzyme-controlled toehold regulation for DNA logic circuits[J]. Nanoscale, 2017, 9(46): 18223–18228. doi: 10.1039/C7NR06484E 苏晨, 吉邢虎, 何治柯. 核酸工具酶辅助的信号放大技术在生物分子检测中的应用[J]. 分析科学学报, 2016, 32(2): 273–281. doi: 10.13526/j.issn.1006-6144.2016.02.026SU Chen, JI Xinghu, and HE Zhike. Enzyme assisted signal amplification and its applications in biomolecule detection[J]. Journal of Analytical Science, 2016, 32(2): 273–281. doi: 10.13526/j.issn.1006-6144.2016.02.026 LAKIN M R, YOUSSEF S, POLO F, et al. Visual DSD: A design and analysis tool for DNA strand displacement systems[J]. Bioinformatics, 2011, 27(22): 3211–3213. doi: 10.1093/bioinformatics/btr543 SPACCASASSI C, LAKIN M R, and PHILLIPS A. A logic programming language for computational nucleic acid devices[J]. ACS Synthetic Biology, 2019, 8(7): 1530–1547. doi: 10.1021/acssynbio.8b00229 李娜, 丁宝全, 颜颢. DNA纳米技术与生物编程[J]. 中国科学院院刊, 2014, 29(1): 55–69.LI Na, DING Baoquan, and YAN Hao. DNA nanotechnology and bio-programming[J]. Bulletin of the Chinese Academy of Sciences, 2014, 29(1): 55–69. -





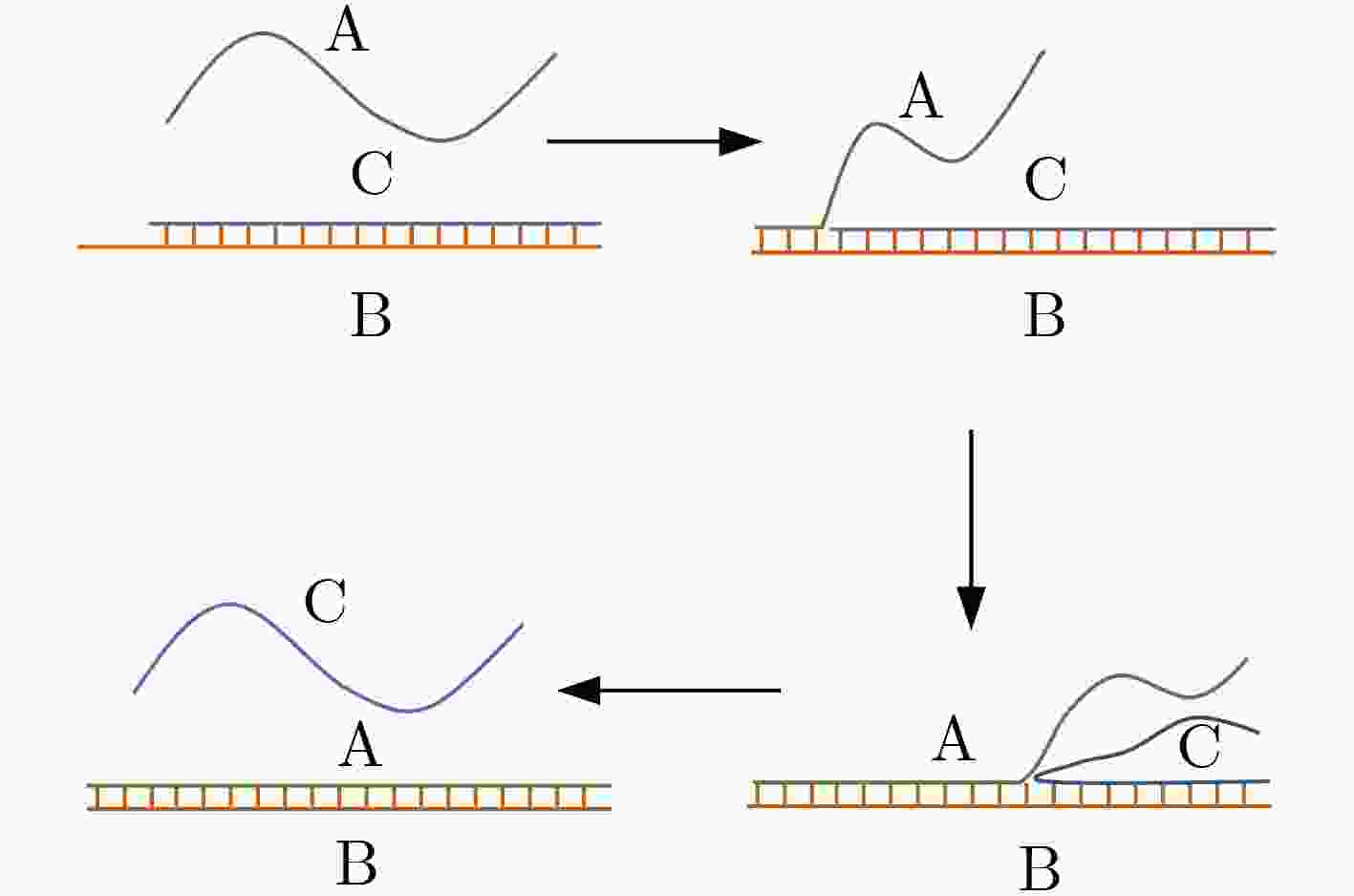
 下载:
下载:
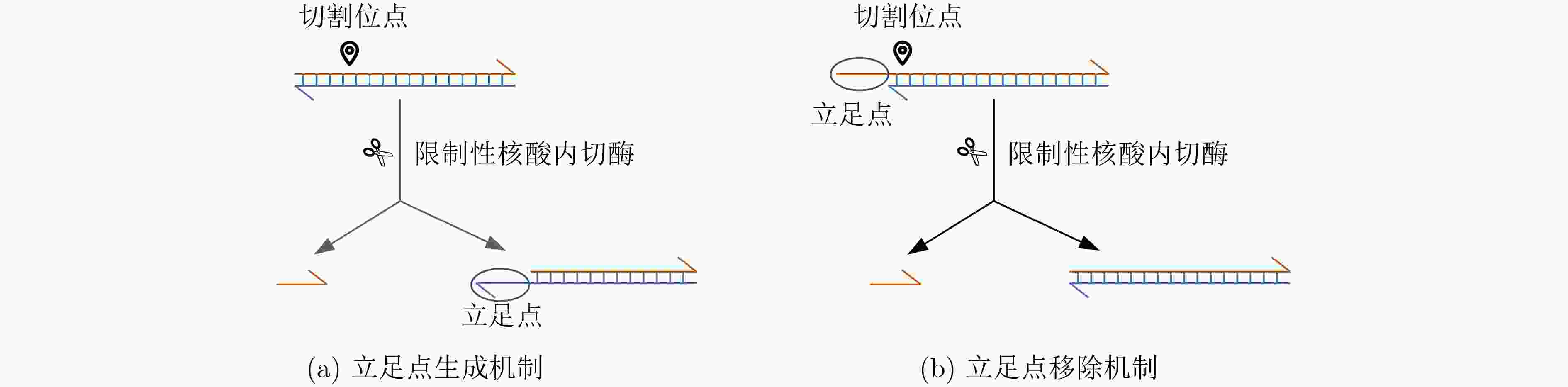
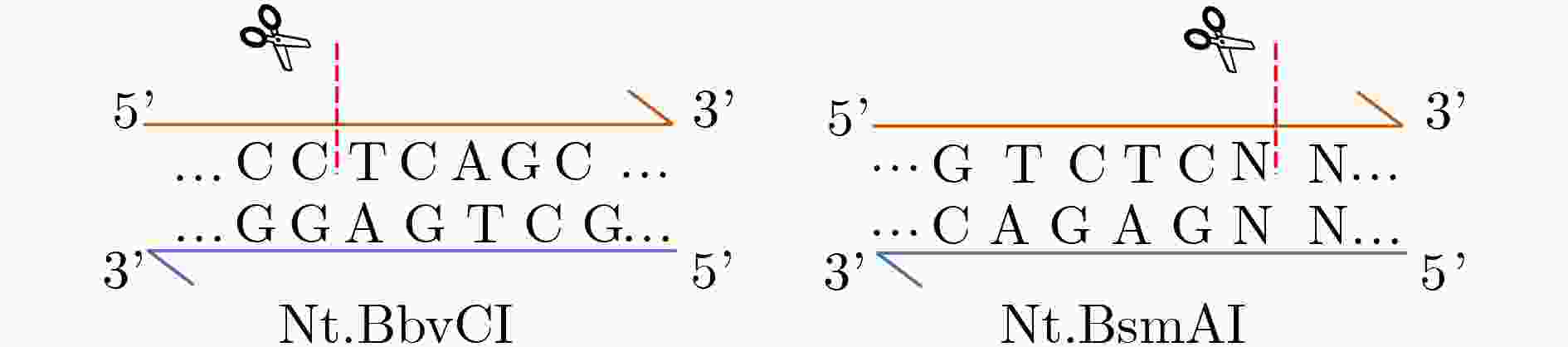
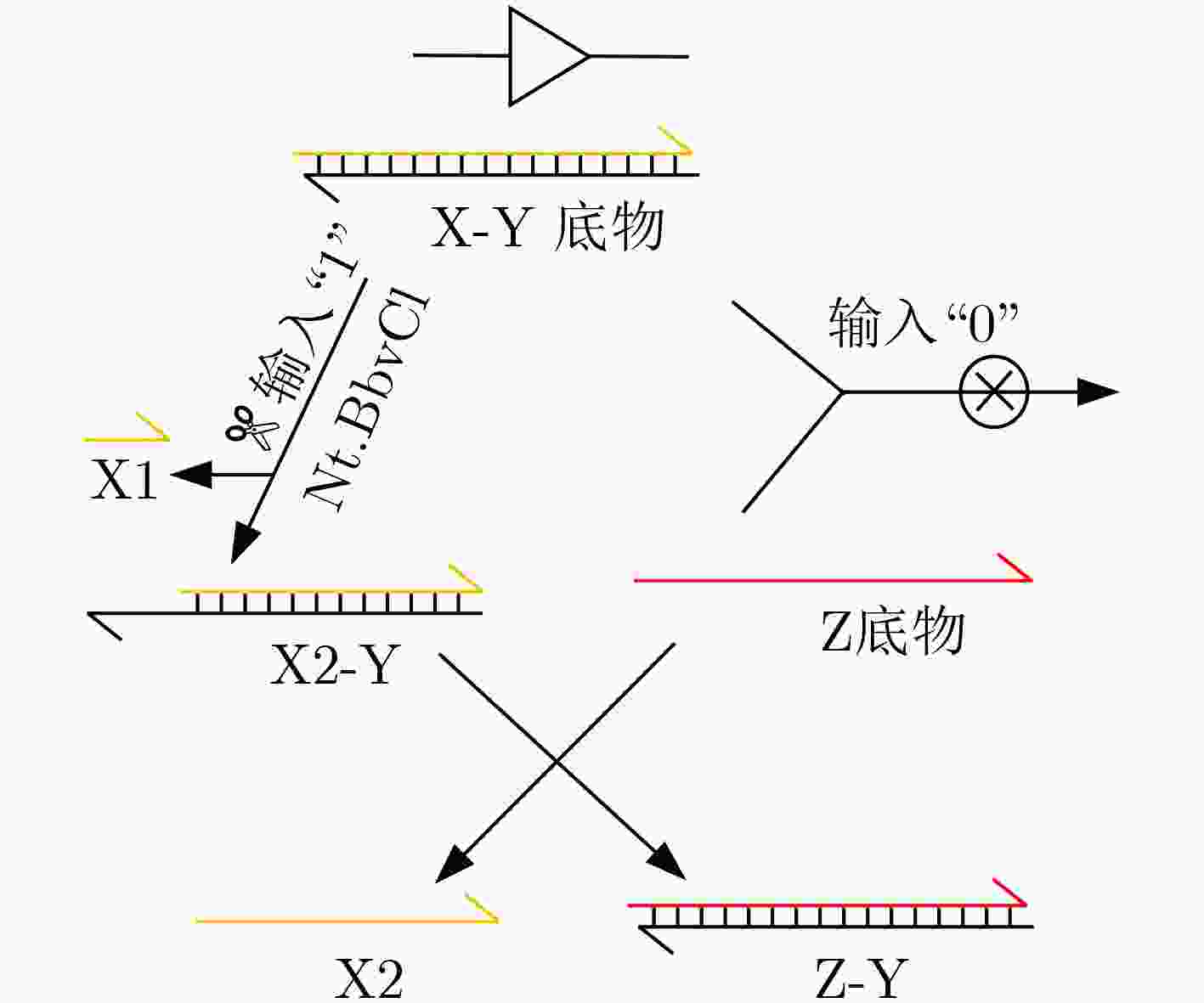
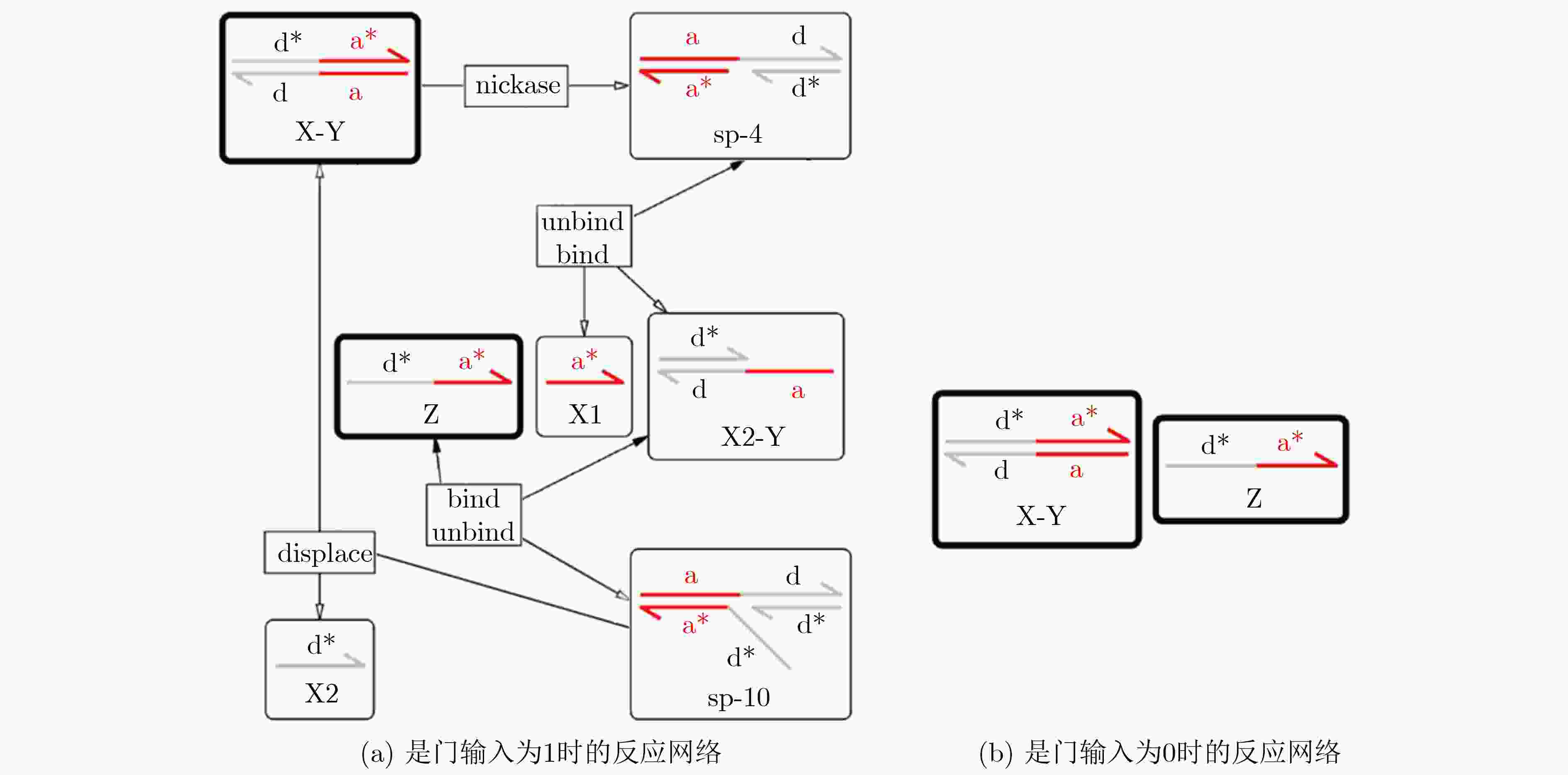
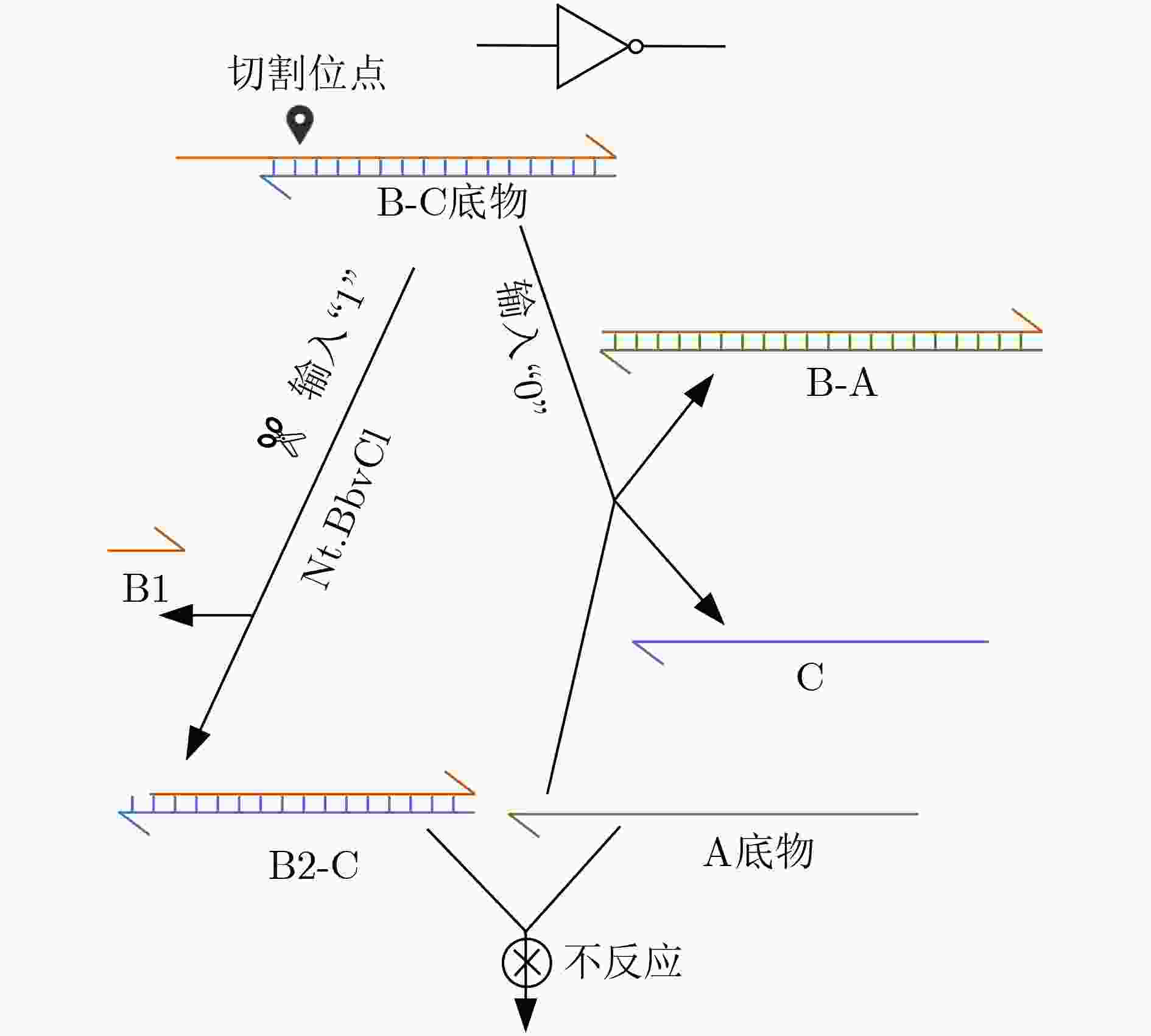
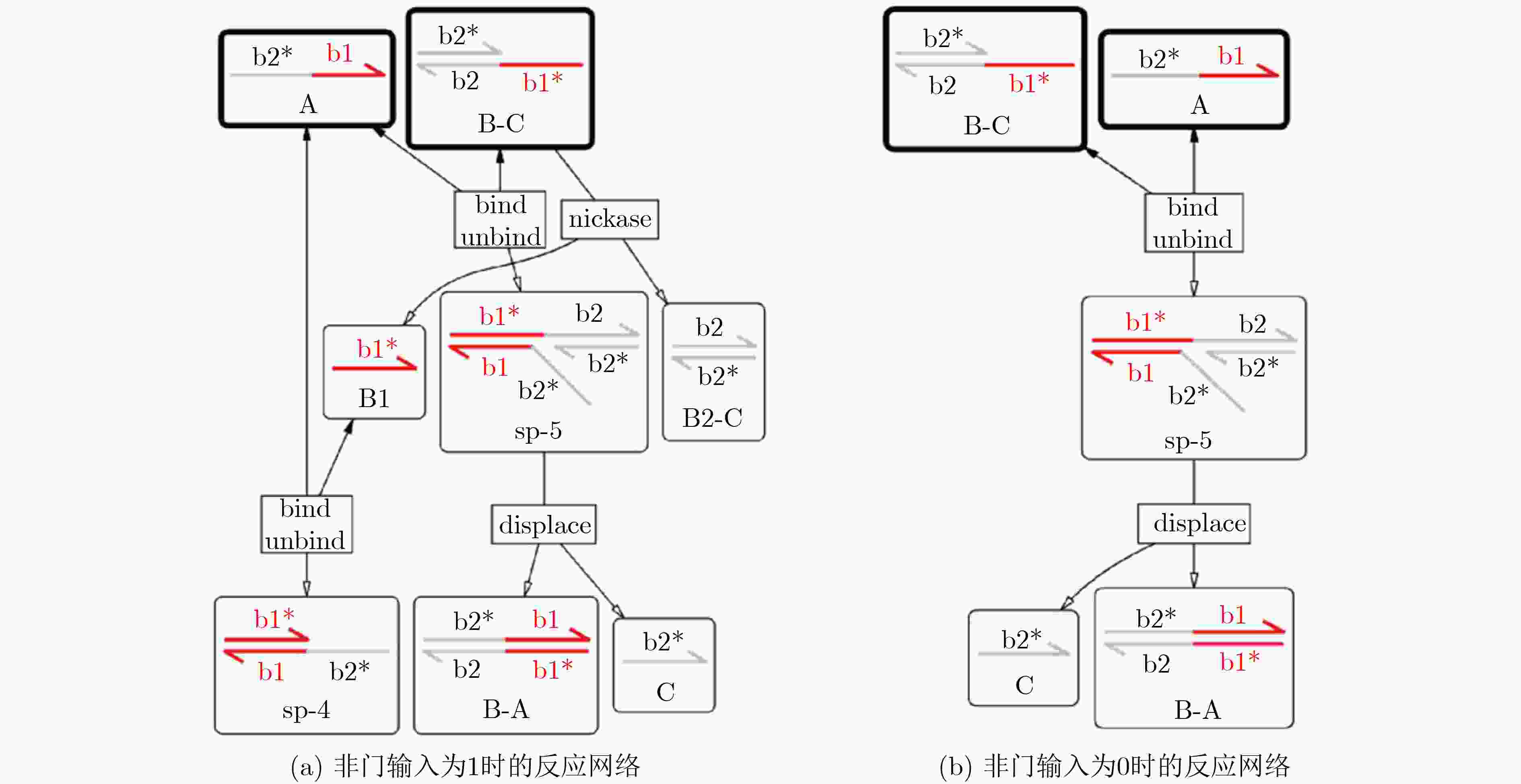
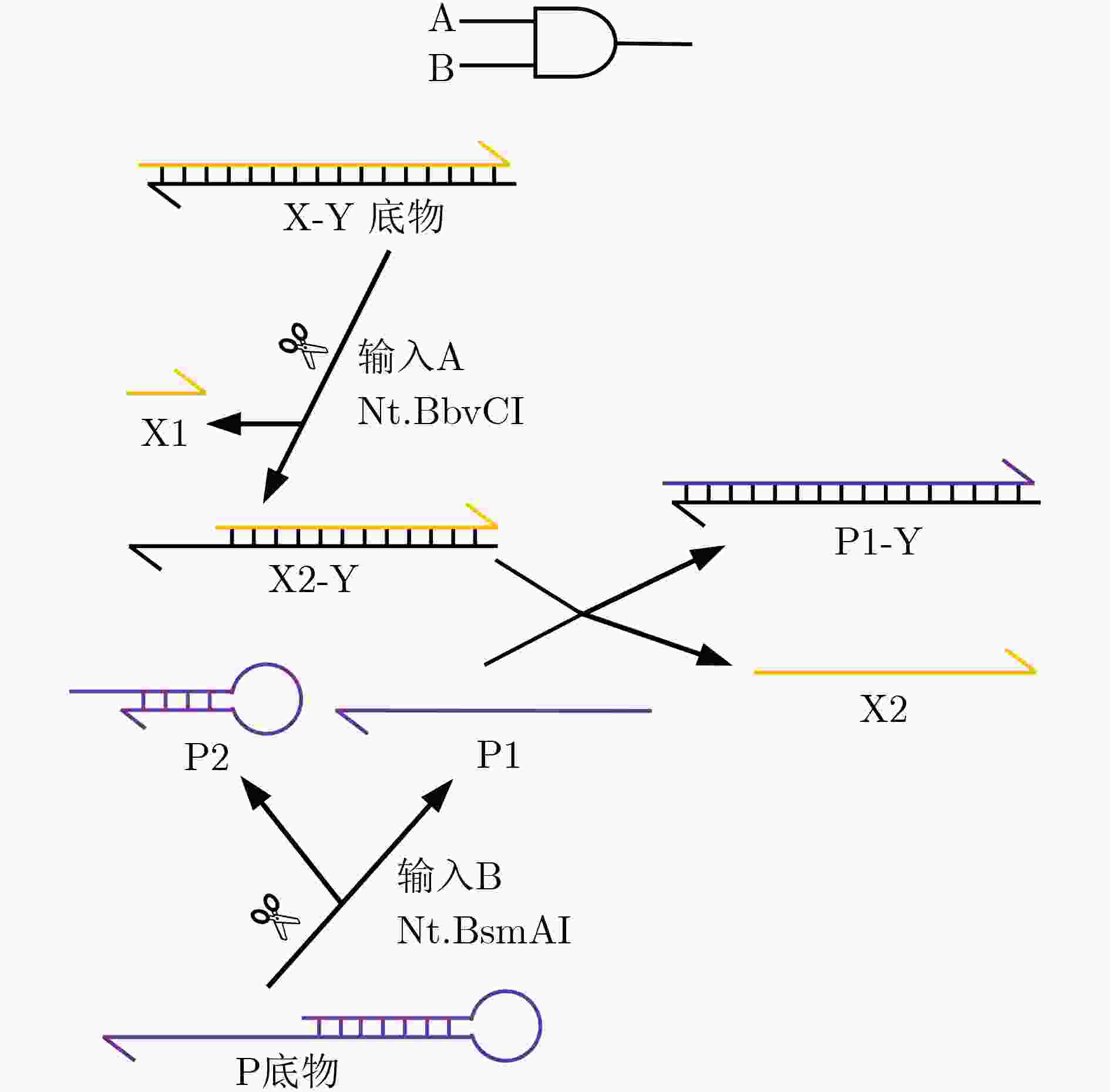
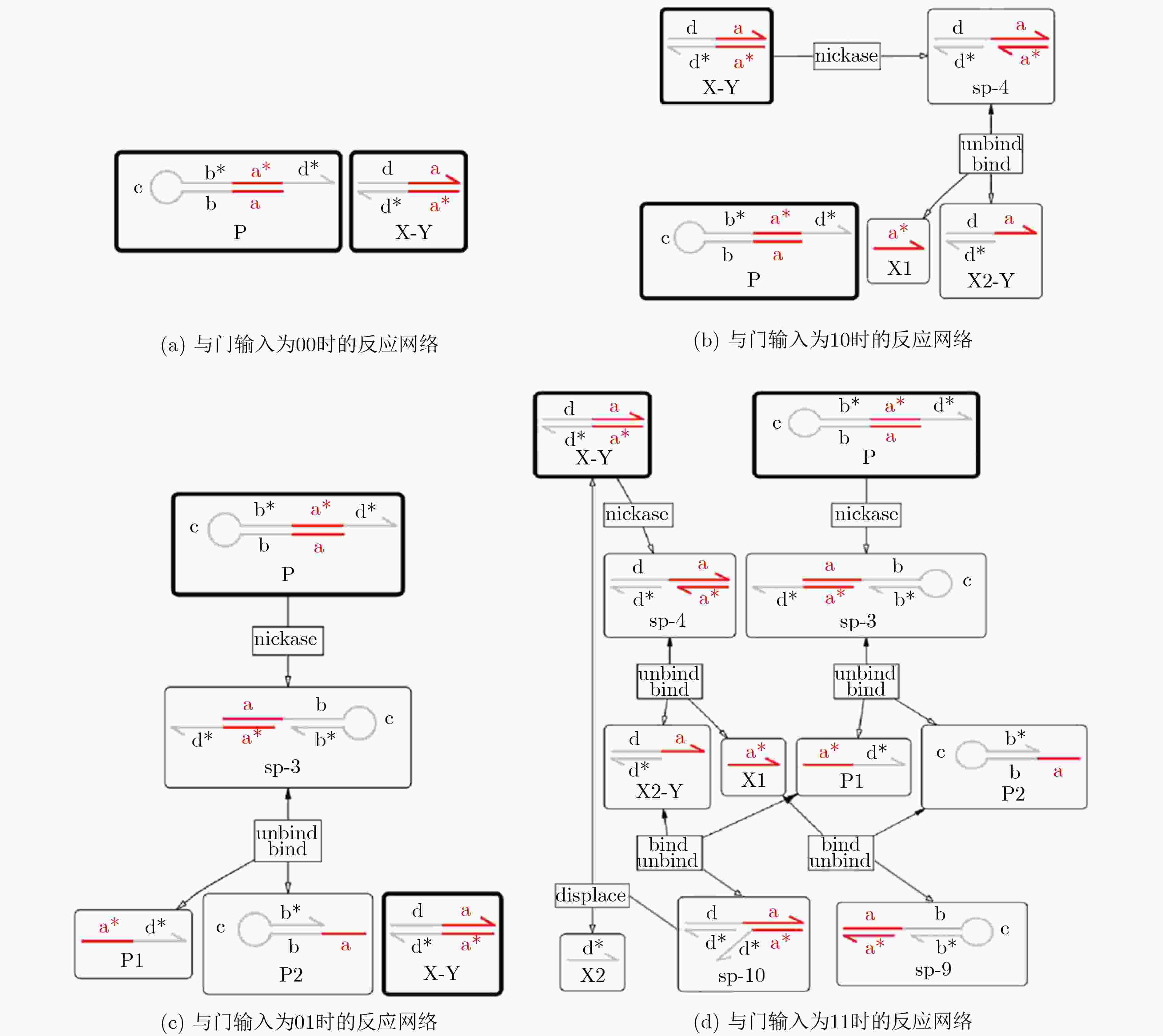
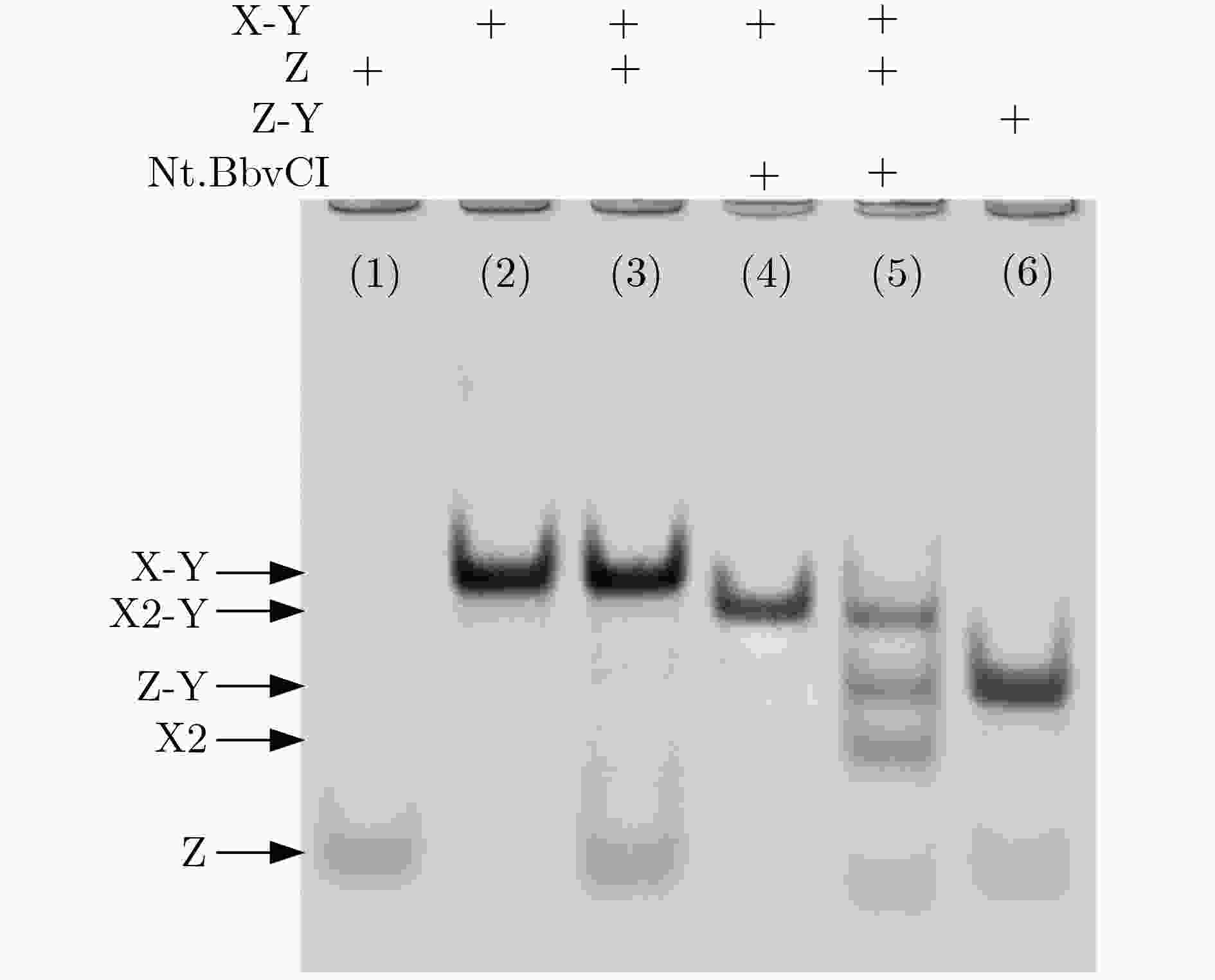
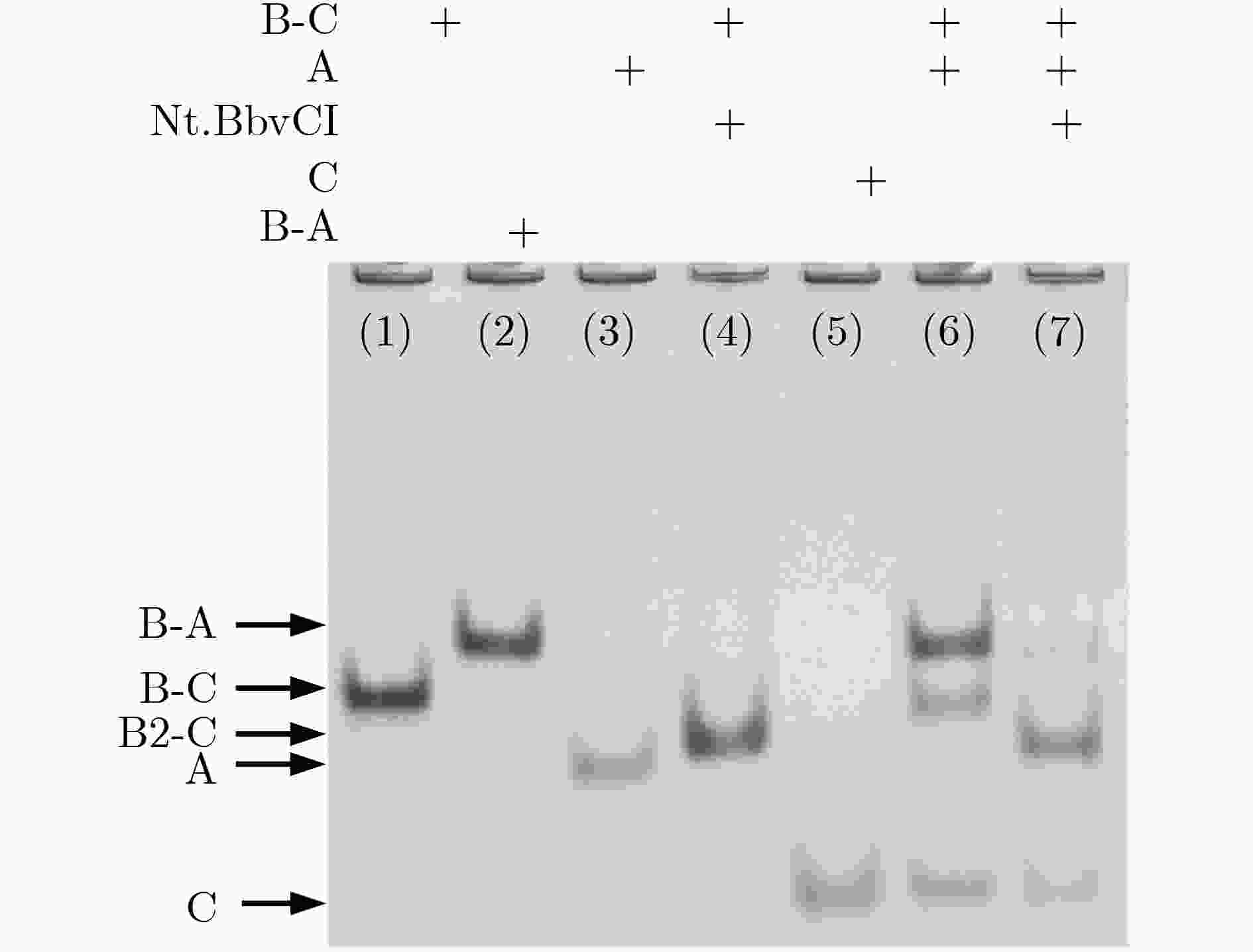
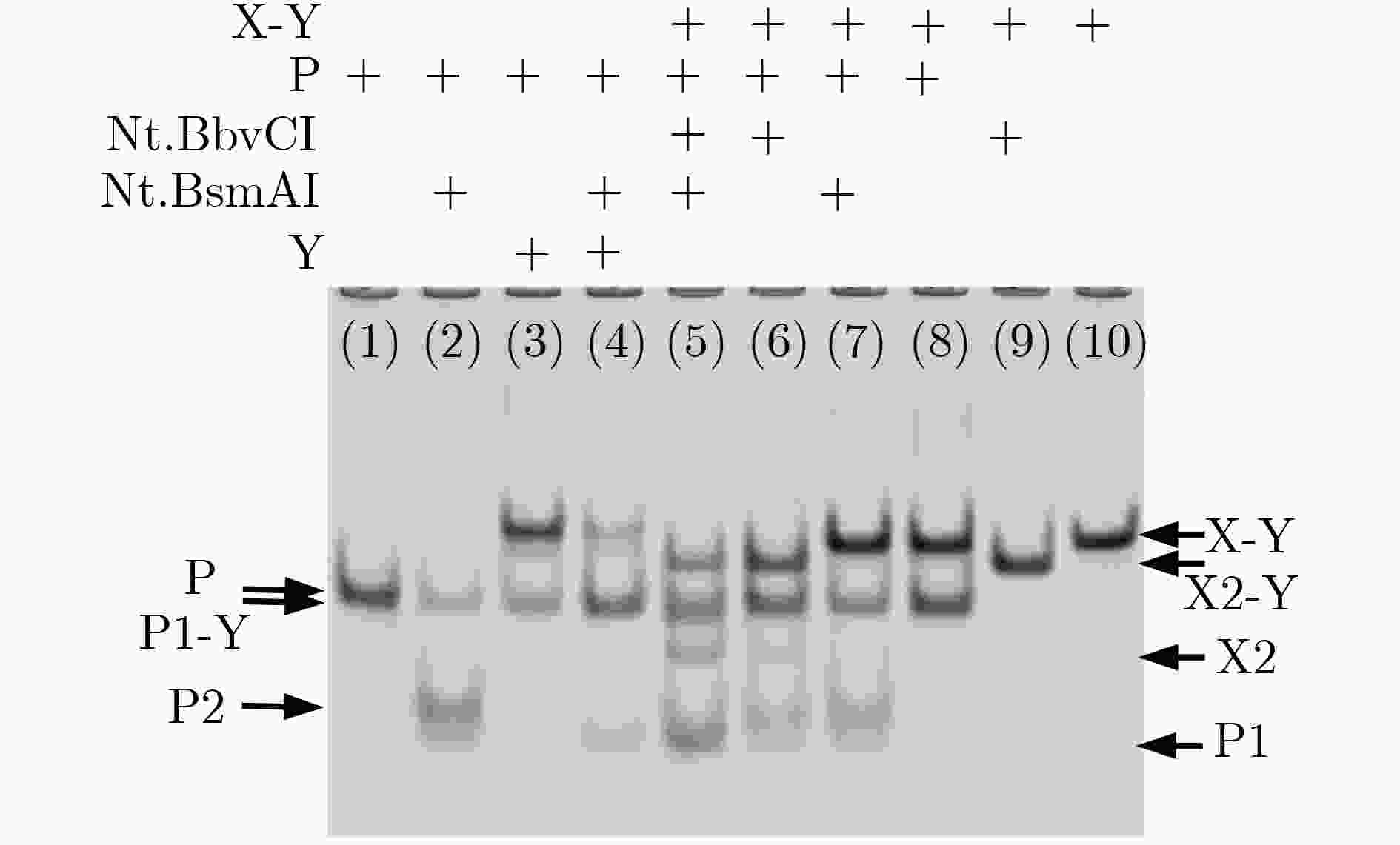


 下载:
下载:
