Multi-context Autoencoders for Multivariate Medical Signals Based on Deep Convolutional Neural Networks
-
摘要:
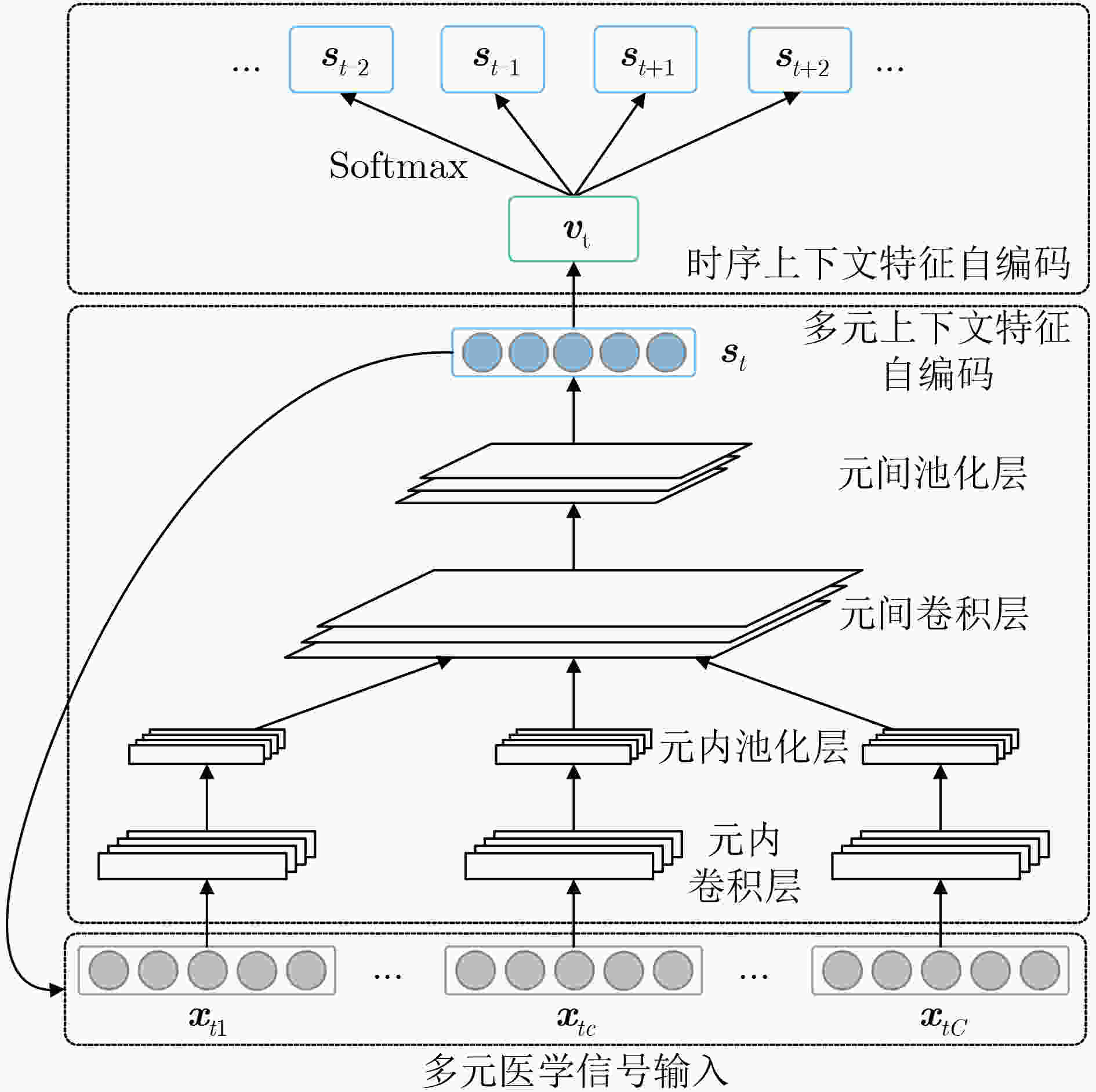
多元医学信号的典型代表有多模态睡眠图和多通道脑电图等,采用无监督深度学习表征多元医学信号是目前健康信息学领域中的一个研究热点。为了解决现有模型没有充分结合医学信号多元时序结构特点的问题,该文提出了一种无监督的多级上下文深度卷积自编码器(mCtx-CAE)。首先改进传统卷积神经网络结构,提出一种多元卷积自编码模块,以提取信号片段内的多元上下文特征;其次,提出采用语义学习技术对信号片段间的时序信息进行自编码,进一步提取时序上下文特征;最后通过共享特征表示设计目标函数,训练端到端的多级上下文自编码器。实验结果表明,该文所提模型在两种应用于不同医疗场景下的多模态和多通道数据集(UCD和CHB-MIT)上表现均优于其它无监督特征学习方法,能有效提高多元医学信号的融合特征表达能力,对提高临床时序数据的分析效率有着重要意义。
Abstract:Learning unsupervised representations from multivariate medical signals, such as multi-modality polysomnography and multi-channel electroencephalogram, has gained increasing attention in health informatics. In order to solve the problem that the existing models do not fully incorporate the characteristics of the multivariate-temporal structure of medical signals, an unsupervised multi-Context deep Convolutional AutoEncoder (mCtx-CAE) is proposed in this paper. Firstly, by modifying traditional convolutional neural networks, a multivariate convolutional autoencoder is proposed to extract multivariate context features within signal segments. Secondly, semantic learning is adopted to auto-encode temporal information among signal segments, to further extract temporal context features. Finally, an end-to-end multi-context autoencoder is trained by designing objective function based on shared feature representation. Experimental results conducted on two public benchmark datasets (UCD and CHB-MIT) show that the proposed model outperforms the state-of-the-art unsupervised feature learning methods in different medical tasks, demonstrating the effectiveness of the learned fusional features in clinical settings.
-
表 1 多元卷积自编码模块具体配置参数
编码单元 卷积层 非线性变换 池化层 元内编码单元 $1 \times 3 \times 16$ ReLU $1 \times 2$ 元间编码单元 $C \times 3 \times 8$ ReLU $1 \times 2$ 解码单元 反卷积层 非线性变换 反池化层 元间解码单元 $C \times 3 \times 8$ ReLU $1 \times 2$ 元内解码单元 $1 \times 3 \times 16$ ReLU $1 \times 2$ 表 2 CHB-MIT数据库上的方法比较结果
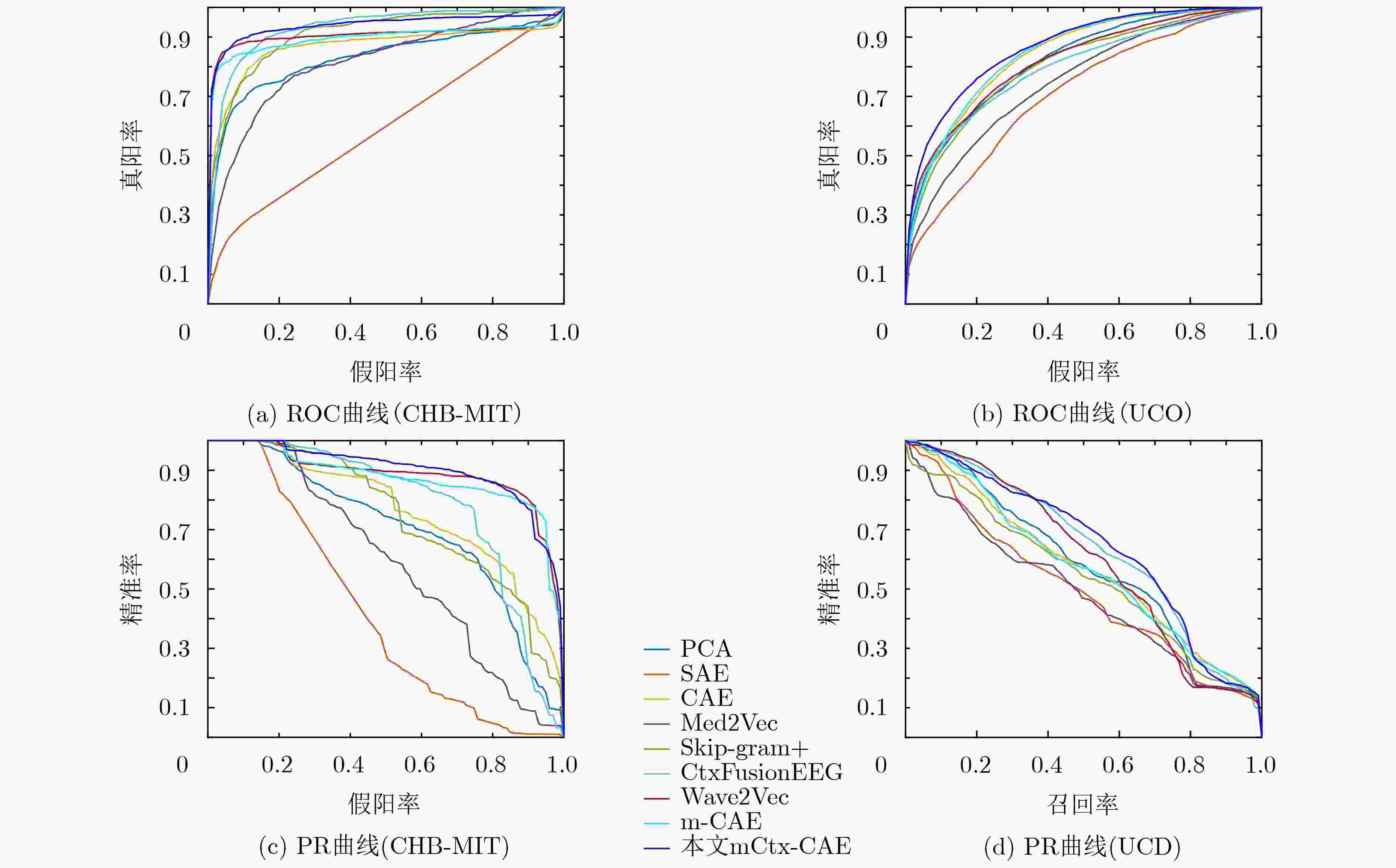
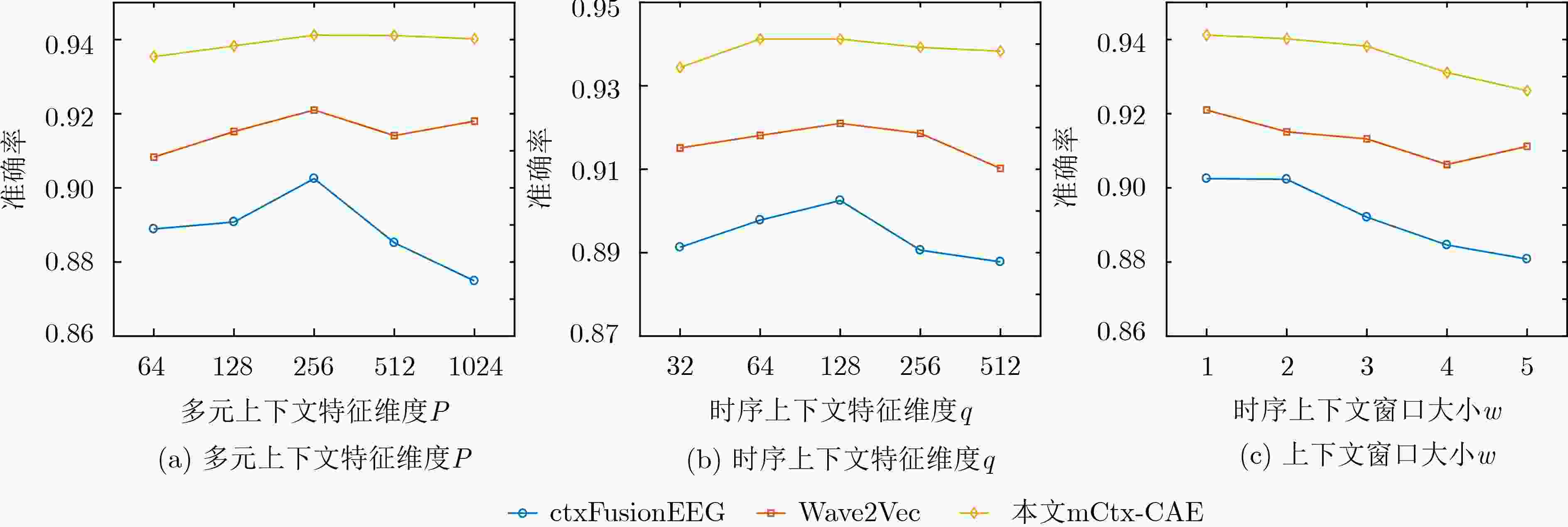
方法 AUC-ROC AUC-PR F1分子 准确率 PCA 0.8291 ± 0.0434 0.7021 ± 0.0872 0.6421 ± 0.0223 0.8768 ± 0.0223 SAE 0.5934 ± 0.0377 0.4180 ± 0.1189 0.0668 ± 0.0415 0.7987 ± 0.0309 CAE 0.8657 ± 0.0305 0.7646 ± 0.0881 0.6277 ± 0.1246 0.8690 ± 0.0267 Med2Vec 0.8155 ± 0.1181 0.5870 ± 0.1670 0.6066 ± 0.2363 0.8351 ± 0.0359 Skip-gram+ 0.9090 ± 0.0356 0.7467 ± 0.1540 0.6288 ± 0.2040 0.8898 ± 0.0173 CtxFusionEEG 0.9287 ± 0.0306 0.7833 ± 0.1147 0.7202 ± 0.1485 0.9025 ± 0.0104 Wave2Vec 0.9035 ± 0.0371 0.8839 ± 0.0261 0.8267 ± 0.0184 0.9210 ± 0.0099 m-CAE 0.8946 ± 0.0401 0.8727 ± 0.0189 0.8417 ± 0.0131 0.9324 ± 0.0058 mCtx-CAE 0.9372 ± 0.0495 0.8980 ± 0.0333 0.8493 ± 0.0191 0.9412 ± 0.0110 表 3 UCD数据库上的方法比较结果
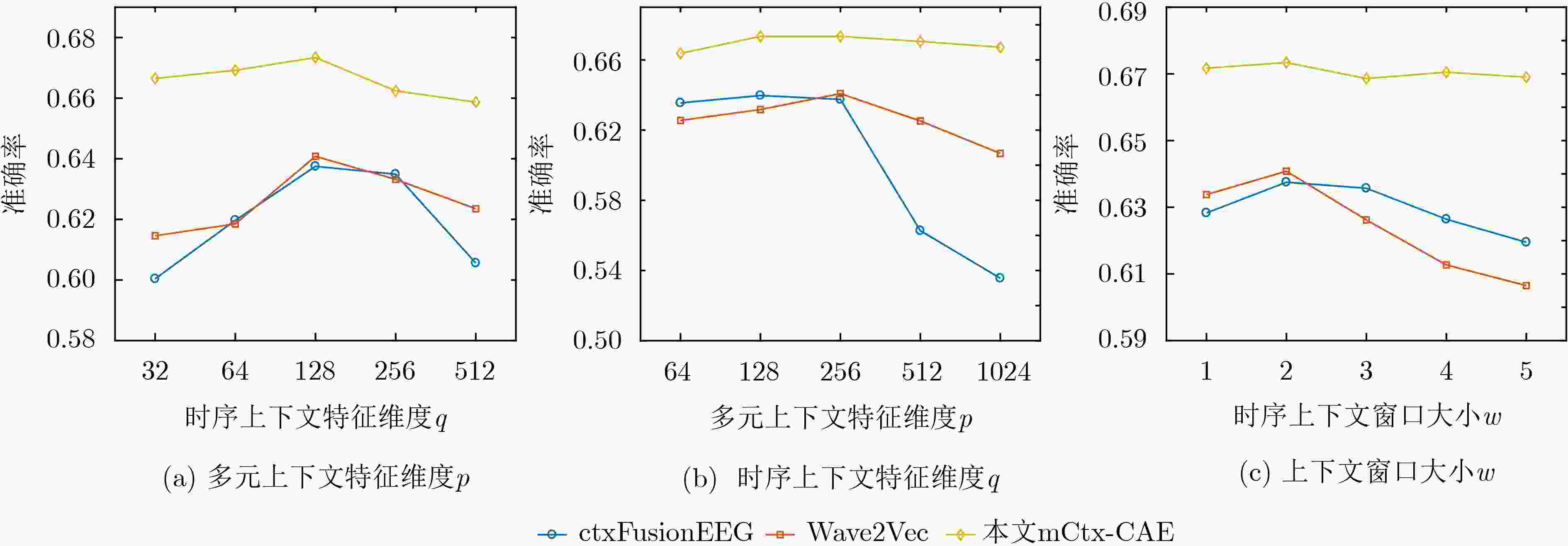
方法 AUC-ROC AUC-PR F1分数 准确率 PCA 0.8177 ± 0.0142 0.5764 ± 0.0172 0.5204 ± 0.0275 0.6193 ± 0.0638 SAE 0.7068 ± 0.1372 0.4965 ± 0.0951 0.2760 ± 0.1815 0.4917 ± 0.1364 CAE 0.8386 ± 0.0376 0.5710 ± 0.0429 0.5180 ± 0.0701 0.6208 ± 0.0961 Med2Vec 0.7479 ± 0.0796 0.4836 ± 0.1046 0.3997 ± 0.1361 0.5619 ± 0.0619 Skip-gram+ 0.8010 ± 0.0992 0.5406 ± 0.0995 0.4342 ± 0.1731 0.5884 ± 0.1077 CtxFusionEEG 0.7941 ± 0.1485 0.6358 ± 0.0709 0.5171 ± 0.1994 0.6375 ± 0.1074 Wave2Vec 0.8161 ± 0.0507 0.5984 ± 0.0698 0.5268 ± 0.0661 0.6408 ± 0.0723 m-CAE 0.8446 ± 0.0361 0.5727 ± 0.0215 0.5600 ± 0.0482 0.6562 ± 0.0767 mCtx-CAE 0.8648 ± 0.0258 0.6423 ± 0.0452 0.5655 ± 0.0228 0.6734 ± 0.0562 -
JOHNSON A E W, GHASSEMI M M, NEMATI S, et al. Machine learning and decision support in critical care[J]. Proceedings of the IEEE, 2016, 104(2): 444–466. doi: 10.1109/JPROC.2015.2501978 RAVI D, WONG C, DELIGIANNI F, et al. Deep learning for health informatics[J]. IEEE Journal of Biomedical and Health Informatics, 2017, 21(1): 4–21. doi: 10.1109/JBHI.2016.2636665 BOOSTANI R, KARIMZADEH F, and NAMI M. A comparative review on sleep stage classification methods in patients and healthy individuals[J]. Computer Methods and Programs in Biomedicine, 2017, 140: 77–91. doi: 10.1016/j.cmpb.2016.12.004 YUAN Ye, XUN Guangxu, JIA Kebin, et al. A multi-view deep learning framework for EEG seizure detection[J]. IEEE Journal of Biomedical and Health Informatics, 2019, 23(1): 83–94. doi: 10.1109/JBHI.2018.2871678 ACAR E, LEVIN-SCHWARTZ Y, CALHOUN V D, et al. Tensor-based fusion of EEG and fMRI to understand neurological changes in schizophrenia[C]. 2017 IEEE International Symposium on Circuits and Systems, Baltimore, USA, 2017: 1–4. JIA Xiaowei, LI Kang, LI Xiaoyi, et al. A novel semi-supervised deep learning framework for affective state recognition on EEG signals[C]. 2014 IEEE International Conference on Bioinformatics and Bioengineering, Boca Raton, USA, 2014: 30–37. LÄNGKVIST M, KARLSSON L, and LOUTFI A. A review of unsupervised feature learning and deep learning for time-series modeling[J]. Pattern Recognition Letters, 2014, 42: 11–24. doi: 10.1016/j.patrec.2014.01.008 HOLZINGER A. Machine Learning for Health Informatics[M]. Cham: Springer, 2016: 161–182. SUPRATAK A, LI Ling, and GUO Yike. Feature extraction with stacked autoencoders for epileptic seizure detection[C]. The 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, USA, 2014: 4184–4187. YAN Bo, WANG Yong, LI Yuheng, et al. An EEG signal classification method based on sparse auto-encoders and support vector machine[C]. 2016 IEEE/CIC International Conference on Communications in China, Chengdu, China, 2016: 1–6. LIN Qin, YE Shuqun, HUANG Xiumei, et al. Classification of epileptic EEG signals with stacked sparse autoencoder based on deep learning[C]. The 12th International Conference on Intelligent Computing, Lanzhou, China, 2016: 802–810. YANG Jianli, BAI Yang, LI Guojun, et al. A novel method of diagnosing premature ventricular contraction based on sparse auto-encoder and softmax regression[J]. Bio-medical Materials and Engineering, 2015, 26(S1): S1549–S1558. doi: 10.3233/BME-151454 XUN Guangxu, JIA Xiaowei, and ZHANG Aidong. Detecting epileptic seizures with electroencephalogram via a context-learning model[J]. BMC Medical Informatics and Decision Making, 2016, 16(Suppl 2): 70. doi: 10.1186/s12911-016-0310-7 LI Xiaoyi, JIA Xiaowei, XUN Guangxu, et al. Improving EEG feature learning via synchronized facial video[C]. 2015 IEEE International Conference on Big Data, Santa Clara, USA, 2015: 843–848. YUAN Ye, XUN Guangxu, SUO Qiuling, et al. Wave2Vec: Deep representation learning for clinical temporal data[J]. Neurocomputing, 2019, 324: 31–42. doi: 10.1016/j.neucom.2018.03.074 YUAN Ye, XUN Guangxu, JIA Kebin, et al. A multi-context learning approach for EEG epileptic seizure detection[J]. BMC Systems Biology, 2018, 12(6): 47–57. doi: 10.1186/s12918-018-0626-2 ZHANG Junming, WU Yan, BAI Jing, et al. Automatic sleep stage classification based on sparse deep belief net and combination of multiple classifiers[J]. Transactions of the Institute of Measurement and Control, 2016, 38(4): 435–451. doi: 10.1177/0142331215587568 YULITA I N, FANANY M I, and ARYMURTHY A M. Sequence-based sleep stage classification using conditional neural fields[J]. arXiv preprint arXiv:1610.01935 , 2016. LÄNGKVIST M, KARLSSON L, and LOUTFI A. Sleep stage classification using unsupervised feature learning[J]. Advances in Artificial Neural Systems, 2012, 2012: 107046. doi: 10.1155/2012/107046 MASCI J, MEIER U, CIREŞAN D, et al. Stacked convolutional auto-encoders for hierarchical feature extraction[C]. The 21st International Conference on Artificial Neural Networks, Espoo, Finland, 2011: 52–59. HINTON G E and SALAKHUTDINOV R R. Reducing the dimensionality of data with neural networks[J]. Science, 2006, 313(5786): 504–507. doi: 10.1126/science.1127647 MIKOLOV T, SUTSKEVER I, CHEN Kai, et al. Distributed representations of words and phrases and their compositionality[C]. The 26th International Conference on Neural Information Processing Systems, Lake Tahoe, USA, 2013: 3111–3119. CHOI E, BAHADORI M T, SEARLES E, et al. Multi-layer representation learning for medical concepts[C]. The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, USA, 2016: 1495–1504. SHOEB A H. Application of machine learning to epileptic seizure onset detection and treatment[D]. [Ph.D. dissertation], Massachusetts Institute of Technology, 2009. GOLDBERGER A L, AMARAL L A N, GLASS L, et al. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals[J]. Circulation, 2000, 101(23): E215–E220. doi: 10.1161/01.CIR.101.23.e215 FAWCETT T. An introduction to ROC analysis[J]. Pattern Recognition Letters, 2006, 27(8): 861–874. doi: 10.1016/j.patrec.2005.10.010 DAVIS J and GOADRICH M. The relationship between precision-recall and ROC curves[C]. The 23rd International Conference on Machine Learning, Pittsburgh, USA, 2006: 233–240. HE Haibo and GARCIA E A. Learning from imbalanced data[J]. IEEE Transactions on Knowledge and Data Engineering, 2009, 21(9): 1263–1284. doi: 10.1109/TKDE.2008.239 ZEILER M D. ADADELTA: An adaptive learning rate method[J]. arXiv preprint arXiv:1212.5701, 2012. -





 下载:
下载:


 下载:
下载:
